Abstract
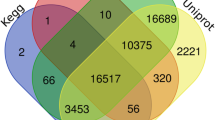
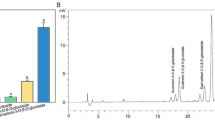
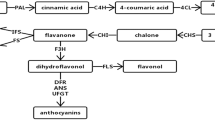
Toona sinensis, a member of Meliaceae family, is a traditional Chinese woody vegetable widely used as food and in health since ancient times. T. sinensis bud has extensive clinical uses because of its high flavonoid content. However, the literature lacks information on flavonoid metabolism and characterization of the corresponding genes in T. sinensis. In this study, we constructed two cDNA libraries of green (GYC-2) and purple toons (BYC-2) distributed in Taihe County of Anhui Province in China. A total of 9.48 Gb of raw sequencing reads were generated using Illumina technology. Obtained raw reads were assembled into 66,331 non-redundant unigenes with mean length 1076 bp. A total of 50,582 unigenes (accounting for 76. 26% of all-unigenes) could be matched to public database using BLASTx. Through alignment against KEGG database, a total of 34,183 were annotated into 135 KEGG pathways. Among such pathways, many candidate genes that are associated with flavonoid biosynthesis were discovered in our transcriptome data. In total, 9541 unigenes identified were differentially expressed genes (DEGs), including 5408 up-regulated unigenes and 4133 down-regulated unigenes in green toon vs purple toon. Moreover, numerous transcription factors (MYB, bHLH, and WD40) were found in our data. Many genes related to flavonoid biosynthesis showed preferential expression in BYC-2 cultivar. Therefore, de novo transcriptome analysis of unique transcripts provides an invaluable resource for exploring vital gene related to flavonoid biosynthesis of T. sinensis bud.









Similar content being viewed by others
References
Assefa AD, Ko EY, Moon SH, Keum YS (2016) Antioxidant and antiplatelet activities of flavonoid-rich fractions of three citrus fruits from Korea. 3 Biotech 6:109
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7:986–995
Boss PK, Davies C, Robinson SP (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv shiraz grape berries and the implications for pathway regulation. Plant Physiol 111:1059–1066
Conesa A, Cötz S, García-Gómez JM, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Dixon RA, Liu CG, Jun JH (2013) Metabolic engineering of anthocyanins and condensed tannins in plants. Curr Opin Biotechnol 24(2):329–335
Galli V, Guzman F, Messias RS, Körbes AP, Silva SDA, Margis-Pinheiro M, Margis R (2014) Transcriptome of tung tree mature seeds with an emphasis on lipid metabolism genes. Tree Genet Genomes 10:1353–1367
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:761–780
Hsu CY, Huang PL, Chen CM, Mao CT, Chaw SM (2012) Tangy Scent in Toona sinensis (Meliaceae) leaflets: isolation, functional characterization, and regulation of TsTPS1 and TsTPS2, two key terpene synthase genes in the biosynthesis of the scent compound. Curr Pharm Biotechnol 13:1–12
Huang PJ, Hseu YC, Lee MS, Kumar KJS, Wu CR, Hsu LS, Liao JW, Cheng IS, Kuo YT, Huang SY, Yang HL (2012) In vitro and in vivo activity of gallic acid and Toona sinensis leaf extracts against HL-60 human premyelocytic leukemia. Food Chem Toxicol 50(10):3489–3497
Huo JW, Liu P, Wang Y, Qin D, Zhao LJ (2016) De novo transcriptome sequencing of blue honeysuckle fruit (Lonicera caerulea L.) and analysis of major genes involved in anthocyanin biosynthesis. Acta Physiol Plant 38:180
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18(9):477–483
Jin B, Dong HR (1994) Nutritious substances and pigments in Chinese toon sprouts and physiological changes during storage. Acta Agric Shanghai 10(4):84–88
Kakumu A, Ninomiya M, Efdi M, Adfa M, Hayashi M, Tanaka K, Koketsu M (2014) Phytochemical analysis and antileukemic activity of polyphenolic constituents of Toona sinensis. Bioorg Med Chem Lett 24:4286–4290
Kim KI, van de Wiel MA (2008) Effects of dependence in high-dimensional multiple testing problems. BMC Bioinform 9:114
Liu J, Sun ZX, Chen YT, Jiang JM (2012) Isolation and characterization of microsatellite loci from an endangered tree species. Toona ciliata var. pubescens. Genet Mol Res 11(4):4411–4417
Liu SS, Chen J, Li SC, Zeng X, Meng ZX, Guo SX (2015) Comparative transcriptome analysis of genes involved in GA-GID1-DELLA regulatory module in symbiotic and asymbiotic seed germination of Anoectochilus roxburghii (Wall.) Lindl. (Orchidaceae). Int J Mol Sci 16(12):30190–30203
Mano H, Ogasawara F, Sato K, Higo H, Minobe Y (2007) Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol 143:1252–1268
McMullen MD, Snook M, Lee EA, Byrne PF, Kross H, Musket TA, Houchins K, Coe EH Jr (2001) The biological basis of epistasis between quantitative trait loci for flavone and 3-deoxyanthocyanin synthesis in maize (Zea mays L.). Genome 44:667–676
Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138:1083–1096
Mu RM, Wang XR, Liu SX, Yuan XL, Wang SB, Fan ZQ (2007) Rapid determination of volatile compounds in Toona sinensis (A. Juss.) Roem. by MAE-HS-SPME followed by GC–MS. Chromatographia 65:463–467
Pan ZY, Li Y, Deng XX, Xiao SY (2014) Non-targeted metabolomic analysis of orange (Citrus sinensis [L.] Osbeck) wild type and bud mutant fruits by direct analysis in real-time and HPLC-electrospray mass spectrometry. Metabolomics 10(3):508–523
Ravaglia D, Espley RV, Henry-Kirk RA, Andreotti C, Ziosi V, Hellens RP, Costa G, Allan AC (2013) Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol 13:68
Reuter JA, Spacek DV, Snyder MP (2015) High-throughput sequencing technologies. Mol Cell 58(4):586–597
Routaboul JM, Kerhoas L, Debeaujon I, Pource L, Caboche M, Einhorn J, Lepiniec L (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224(1):96–107
Shi CY, Yang H, Wei CL, Yu O, Zhang ZZ, Jiang CJ, Sun J, Li YY, Chen Q, Xia T, Wan XC (2011) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genom 12:131
Shi SG, Yang M, Zhang M, Wang P, Kang YX, Liu JJ (2014) Genome-wide transcriptome analysis of genes involved in flavonoid biosynthesis between red and white strains of Magnolia sprengeri pamp. BMC Genom 15:706
Shimizu Y, Maeda K, Kato M, Shimomura K (2011) Co-expression of GbMYB1 and GbMYC1 induces anthocyanin accumulation in roots of cultured Gynura bicolor DC. plantlet on methyl jasmonate treatment. Plant Physiol Biochem 49:159–167
Vinodhini V, Lokeswari TS (2014) Antioxidant activity of the isolated compounds, methanolic and hexane extracts of Toona ciliata leaves. Int J Eng Technol 4(3):135–138
Wang KJ, Yang CR, Zhang YJ (2007) Phenolic antioxidants from Chinese toon (fresh young leaves and shoots of Toona sinensis). Food Chem 101(1):365–371
Wang CL, Cao JW, Tian SR, Wang YR, Chen ZQ, Chen MH, Gong GL (2008) Germplasm resources research of Toona sinensis with RAPD and isoenzyme analysis. Biologia 63(3):320–326
Wang XW, Luan JB, Li JM, Bao YY, Zhang CX, Liu SS (2010) De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genom 11:400
Wang X, Li ST, Li J, Li CF, Zhang YS (2015) De novo transcriptome sequencing in Pueraria lobata to identify putative genes involved in isoflavones biosynthesis. Plant Cell Rep 34(5):733–743
Wang JH, Liu JJ, Chen KL, Li HW, He J, Guan B, He L (2016) Anthocyanin biosynthesis regulation in the fruit of Citrus sinensis cv. Tarocco. Plant Mol Biol Rep 34(6):1043–1055
Wu ZJ, Li XH, Liu ZW, Xu ZS, Zhuang J (2014) De novo assembly and transcriptome characterization: novel insights into catechins biosynthesis in Camellia sinensis. BMC Plant Biol 14:277
Xu Q, Chen LL, Ruan XA, Chen DJ, Zhu AD, Chen CL, Bertrand D, Jiao WB, Hao BH, Lyon MP, Chen JJ, Gao S, Xing F, Lan H, Chang JW, Ge XH, Lei Y, Hu Q, Miao Y, Wang L, Xiao SX, Biswas MK, Zeng WF, Guo F, Cao HB, Yang XM, Xu XW, Cheng YJ, Xu J, Liu JH, Luo OJH, Tang ZH, Guo WW, Kuang HH, Zhang HY, Roose ML, Nagarajan N, Deng XX, Ruan YJ (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45(1):59–66
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20:176–185
Yang JX, Yang Y, Li WY, Qu CQ (2010) Determination of quercetin in Taihe Toona Sinensis by high performance liquid chromatography. Pharm Biotechnol 17(6):513–515
Yang SJ, Zhao Q, Xiang HM, Liu MJ, Zhang QY, Xue W, Song BA, Yang S (2013) Antiproliferative activity and apoptosis-inducing mechanism of constituents from Toona sinensis on human cancer cells. Cancer Cell Int 13:12
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:293–297
Yu KQ, Xu Q, Da XL, Guo F, Ding YD, Deng XX (2012) Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). BMC Genom 13:10
Zhang W, Li C, You LJ, Fu X, Chen YS, Luo YQ (2014) Structural identification of compounds from Toona sinensis leaves with antioxidant and anticancer activities. J Funct Foods 10:427–435
Zhang MF, Jiang LM, Zhang DM, Jia GX (2015) De novo transcriptome characterization of Lilium ‘Sorbonne’ and key enzymes related to the flavonoid biosynthesis. Mol Genet Genom 290:399–412
Zhang X, Song ZQ, Liu T, Guo LL, Li XF (2016) De Novo assembly and comparative transcriptome analysis provide insight into lysine biosynthesis in Toona sinensis Roem. Int J Genom 3:1–9
Zhou GX, Zhang BG, Lin L, Zhu Q, Guo L, Pu YY, Cao X (2010) Study on the relationship between Toona sinensis Roem stand productivity and site conditions in Sichuan Basin. Ecol Econ 6:387–394
Zhou XJ, Dong Y, Zhao JJ, Huang L, Ren XP, Chen YN, Huang SM, Liao BS, Lei Y, Yan LY, Jiang HF (2016) Genomic survey sequencing for development and validation of single-locus SSR markers in peanut (Arachis hypogaea L.). BMC Genom 17:420
Acknowledgements
This research was supported by a grant from the Natural Science Key Foundations of the Anhui Bureau of Education (Nos. KJ2016A552, KJ2015KJ006, and 2014KJ019) and the Innovation Program for College Students (No. 201510371061).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. H. Walter.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, H., Ren, L., Fan, X. et al. Identification of putative flavonoid-biosynthetic genes through transcriptome analysis of Taihe Toona sinensis bud. Acta Physiol Plant 39, 122 (2017). https://doi.org/10.1007/s11738-017-2422-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-017-2422-9