Abstract
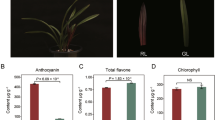
Blue honeysuckle (Lonicera caerulea L.) is an economically important berry because its flavonoid and anthocyanin contents are generally higher than those of other fruits. However, research on anthocyanin accumulation in blue honeysuckle is limited by insufficient genome information. In this paper, Illumina HiSeq™ 2000 platform was used for the transcriptome sequencing of blue honeysuckle fruit in two periods (green-ripe and veraison periods). This platform was also used to obtain more than 116 million sequencing reads. Those reads were de novo assembled and produced 66,610 All-unigenes. After a comparison between currently available public databases (NR, Swiss-prot, KEGG, COG, and GO) and obtained annotated unigenes, 43,202 unigenes (64.86 %) showed similarity to known protein sequences (E value ≤0.00001). The KEGG pathway indicated that some genes related to anthocyanin biosynthesis existed mainly in the secondary metabolic pathway of the samples. A series of enzymes related to anthocyanin biosynthesis was identified in NR annotation; some candidate genes and their metabolic pathways were also found. We conducted a preliminary analysis on the expression of some important enzymes (PAL, CHS, F3H, ANS, and DFR) and transcription factors (MYB, bHLH, and WD40) related to anthocyanin biosynthesis through quantitative real-time PCR to explore the biosynthesis process of anthocyanin and the accuracy of sequencing results. Information on blue honeysuckle transcriptome can serve as an important reference for breeding and production.








Similar content being viewed by others
References
Akada S, Dube SK (1995) Organization of soybean chalcone synthase gene clusters and characterization of a new member of the family. Plant Mol Biol 29:189–199
Albert NW, Lewis DH, Zhang H (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65:771–784
Barbara K, Anna P, Barbara M, Piotr K, Jacek N, Agnieszka B (2012) Phenolic composition and antioxidant properties of polish blue-berried honeysuckle genotypes by HPLC-DAD-MS, HPLC postcolumn derivatization with ABTS or FC, and TLC with DPPH visualization. J Agric Food Chem 60:1755–1763
Baudry A, Heim MA, Dubreucq B (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39(3):366–380
Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetie pathway. Proc Natl Acad 96:12929–12934
Castellarin SD, Pfeiffer AP, Sivilotti M (2007) Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ 30:1381–1399
Chagné D, Lin-Wang K, Espley RV (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161(1):225–239
Christie PJ, Alfenito MR, Walbot V (1994) Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194:541–549
Deluc L, François B, Chloé M (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140:499–511
Espley RV, Hellens RP, Putterill J (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49(3):414–427
Gonzalez A, Zhao M, Leavitt JM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53(5):814–827
Hichri I, Heppel SC, Pillet J (2010) The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol Plant 3(3):509–523
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Huang H, Hu K, Han KT, Xiang QY, Dai SL (2013) Flower color modification of chrysanthemum by suppression of F3 H and overexpression of the exogenous senecio cruentus F3′5′H gene. PLoS One 8:e74395
Jia L, Clegg MT, Tao J (2004) Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol 134:575–585
Kowalczyk E, Krzesinski P, Kura M (2003) Anthocyanins in medicine. Pol J Pharmacol 55(5):699–702
Lemos MA, Aliyu MM, Hungerford G (2015) Influence of cooking on the levels of bioactive compounds in purple majesty potato observed via chemical and spectroscopic means. Food Chem 173:462–467
Li XY, Sun HY, Pei JB, Dong YY, Wang FW, Chen H, Sun YP, Wang N, Li HY, De Li YD (2012) Novo sequencing and comparative analysis of the blueberry transcriptome to discover putative genes related to antioxidants. Gene 511:54–61
Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie T, Espley R, Hellens R, Allan A (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10(1):50
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 25(4):402–408
Mortazavi A, Williams BA, MeCue K et al (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Moyer RA, Hummer KE, Finn CE et al (2002) Anthocyanins, phenolics, and antioxidant capacity in diverse small fruits: Vaccinium, Rubus, and Ribes. J Agric 50:519–525
Nesi N, Debeaujon I, Jond C (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12(10):1863–1878
Pelletier MK, Murrell JR, Shirley BW (1997) Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in arabidopsis further evidence for differential regulation of “early” and “late” genes. Plant Physiol 113:1437–1445
Qi B, Yang Y, Yin Y, Xu M, Li H De (2014) novo sequencing, assembly, and analysis of the Taxodium ‘Zhongshansa’ roots and shoots transcriptome in response to short-term waterlogging. BMC Plant Biol 14:201
Sarma AD, Sharma R (1999) Anthocyanin-DNA copigmentation complex: mutual protection against oxidative damage. Phytochemistry 52:1313–1318
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc 3(6):1101–1108
Virachnee L, Mary M, George S (2008) Determination of anthocyanins in various cultivars of highbush and rabbiteye blueberries. Food Chem 111(1):249–254
Wang LS, Stoner GD (2008) Anthocyanins and their role in cancer prevention. Cancer Lett 269(2):281–290
Wu H, Zhang Y, Zhang W, Pei X, Zhang C, Jia S, Li W (2015) Transcriptomic analysis of the primary roots of Alhagi sparsifolia in response to water stress. PLoS One 10(3):e0120791
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20:176–185
Zhang L, Ming R, Zhang J, Tao A, Fang P, Qi J (2015) De novo transcriptome sequence and identification of major bast-related genes involved in cellulose biosynthesis in jute (Corchorus capsularis L.). BMC Genomics 16(1):1062
Zimmermann IM, Heim MA, Weisshaar B (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40(1):22–34
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant Nos. 31272130).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Peng.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huo, Jw., Liu, P., Wang, Y. et al. De novo transcriptome sequencing of blue honeysuckle fruit (Lonicera caerulea L.) and analysis of major genes involved in anthocyanin biosynthesis. Acta Physiol Plant 38, 180 (2016). https://doi.org/10.1007/s11738-016-2197-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-016-2197-4