Abstract
Jasmonic acid methyl ester (JAMe) has been recently shown to play a crucial role in many physiological processes. In this paper, we focused on cotyledon senescence in Ipomoea nil and revealed that JAMe and darkness are the main factors stimulating the process examined. What is more, we showed that mefenamic acid (a jasmonate biosynthesis inhibitor) reverses the stimulatory effect of darkness on senescence. In plants growing under dark conditions, stimulation of JASMONIC ACID CARBOXYL METHYLTRANSFERASE (InJMT) expression and, consequently, an increase in JAMe content, have been observed. In turn, the level of jasmonic acid (JA) gradually decreased. Moreover, dark-grown seedlings demonstrated a lower PSII functional activity and a reduced chlorophyll content and autofluorescence. All of these data suggest that JAMe is a signal molecule controlling the senescence of cotyledons in I. nil.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The senescence of cotyledons is the final stage of their development. This process is an integrated response of these organs to age information and other environmental signals, both internal and external, such as stresses of salinity, nutrient limitation or darkness (Bouchard and Yamasaki 2008; Corpas et al. 2009; Du et al. 2014; Ma et al. 2008; Neill et al. 2008; Xuan et al. 2010; Zhao et al. 2009). Although there is no single universal mechanism for regulating senescence, some characteristic stages of that process can be distinguished. Initially, thylakoid membrane structures are damaged, the photosynthetic rate drops as a result of chlorophyll degradation, and the catabolic process stops (Pružinská et al. 2005; Troncoso-Poncea et al. 2013; Wrischer et al. 2009; Zhang et al. 2010). Subsequently, the ER, ribosomes, proteins, lipids and nucleic acids are decomposed. In the final stage of cotyledon senescence, the mitochondria are degraded, oxidative phosphorylation intensity is decreased, and the tonoplast and nuclear membrane are interrupted, all of which lead to irreversible changes in the structures of the cytoplasm and the nucleus, as well as to the accumulation of secondary metabolites (Lim et al. 2007; Peterman and Siedow 1985). Although cotyledon senescence has been studied extensively (Ananieva et al. 2008a, b; Kanazawa et al. 2000; Krul 1974; McKersie et al. 1987; Peterman and Siedow 1985; Rukes and Mulkey 1993; Watanabe et al. 1994), its underlying regulatory network is unclear in comparison to leaf senescence (Lim et al. 2007). Numerous studies have shown that leaf senescence correlates with a progressive decline in the photosynthetic rate, which is accompanied by Rubisco and chlorophyll a/b binding protein (CAB) degradation (Avice and Etienne 2014; Gombert et al. 2006; Lu et al. 2002; Van der Graaff et al. 2006). There are crucial photosynthetic parameters commonly known as senescence physiological markers e.g., chlorophyll content, rate of photosynthesis (Gan 2004). In addition, the transcriptional activity of senescence-associated (SAGs) genes increased in leaves before or during senescence in different species (Breeze et al. 2011; Buchanan-Wollaston et al. 2005). Many SAGs are similar to genes probably involved in the breakdown and mobilization of nutrients, such as proteases, RNases, or glutamine synthetases (Lin et al. 2015). An essential role in coordinating anatomical, physiological and biochemical changes accompanying senescence is attributed to plant hormones, such as cytokinin, ethylene, abscisic acid, auxin and jasmonates (JAs) (Ananieva et al. 2008a; Ellis et al. 2005; Hung and Kao 2004; Jing et al. 2008; Okushima et al. 2005). Hormonal pathways are involved in cotyledon senescence—process that may be age-controlled or stimulated by plant responses to stresses. Such hormones may also affect the synthesis and signaling pathways of other to eventually trigger the expression of stress-responsive genes, which in turn influences cotyledon senescence (Lim et al. 2007).
Lipid-derived JAs arise from the enzymatic oxygenation of 18- and 16-carbon triunsaturated fatty acids. The biosynthesis of these hormones encompasses three cellular compartments: chloroplast, peroxisome and cytosol. The first stage occurs in the chloroplasts, where, thanks to the activity of 13-LOX (lipoxygenase), AOS (allene oxide synthase) and AOC (allene oxide cyclase), the 18:3 or 16:3 acid is oxidized and OPDA (12-oxophytodienoic acid) or dnOPDA (dinor-OPDA) is formed, respectively. Both compounds are transported into peroxisomes and with the participation of OPR3 (12-oxophytodienoate reductase), ACX (acyl-CoA oxidase), MFP (multifunctional protein) and KAT (l-3-ketoacyl-CoA thiolase) are first reduced, and then β-oxidized, which ends in the formation of the (+)-7-iso-jasmonic acid (JA) in the cytosol (Wasternack and Kombrink 2010). Then, JA can be metabolized by esterification into methyl (+)-7-iso-jasmonate (JAMe) or by amino acid conjugation to form (+)-7-iso-jasmonoyl-isoleucine (JA-Ile). The second is considered as the active form of the hormone. In addition, JA can be hydroxylated, sulfated, as well as glucosylated (Acosta and Farmer 2010).
The use of molecular biology techniques in research on the JA signal transduction pathway and regulation of its metabolism has led to the discovery that JA derivatives can be also biologically active in many physiological processes. Studies performed in many plant species show that JAMe, formed in the reaction catalyzed by JASMONIC ACID CARBOXYL METHYLTRANSFERASE (JMT), regulates the sprouting of seeds, participates in proper flower morphogenesis and flower induction, and is involved in responses to stress, which also includes promoting effect on leaf senescence (Ananieva et al. 2007; Kęsy et al. 2011; Maciejewska and Kopcewicz 2002; Maciejewska et al. 2004; Norastehnia et al. 2007; Wang 1998; Wilson et al. 2011).
In our former investigation, we showed that JAMe is an inhibitor of photoperiodic flower induction in model short-day plant (SDP) Ipomoea nil, in which the cotyledons are responsible for the perception of light stimuli (Kęsy et al. 2011; Maciejewska and Kopcewicz 2002; Maciejewska et al. 2004). Bearing in mind the involvement of JAMe in the control of many processes, we conducted physiological experiments on the efficiency of PSII, changes in the jasmonate level, chlorophyll content and autofluorescence. What is more, we performed identification and expression analyses of the InJMT gene participating in JA metabolism accompanying cotyledon senescence.
Materials and methods
Plant material
We prepared plant material (I. nil, Chois cv. Violet; Marutane Seed Co., Kyoto, Japan) according to Frankowski et al. (2009). The seedlings were grown in phytotrones under the conditions described by Wilmowicz et al. (2008). On 6th day of cultivation, half of the seedlings were exposed to darkness (D), whereas the remainder to continuous light (CL). The plants were watered with identical amounts of tap water every 2 days (Supplementary Fig. 1).
Plants growing in darkness were treated with mefenamic acid (MEF), a jasmonate biosynthesis inhibitor, at a concentration of 100 μM in 0.05 % Tween 20 (v/v), whereas plants cultivated under CL were treated with JA or JAMe at a concentration of 100 μM in 0.05 % Tween 20 (v/v). The control plants were treated with 0.05 % Tween 20 (v/v). All solutions were applied by small, soft brushes to the cotyledons (about 50 µL per plant). All treatments were repeated every day at the same time starting from 6th day of cultivation for eight subsequent days. Before each treatment, the cotyledons from darkness and those after MEF application were collected (day 1, 2, 4, 6 and 8). In addition, to monitor the ongoing process of aging, the amount of quantum efficiency of PSII in the cotyledons was simultaneously investigated. The collected material was placed immediately in frozen liquid nitrogen and stored at −80 °C. Each experiment was repeated at least three times (biological repetitions). All data are presented as mean ± standard error (SE).
Determination of chlorophyll content and fluorescence measurement
The chlorophyll content was determined according to Glazińska et al. (2014) with the method using Opti-Sciences CCM-200 (Opti-Sciences, Inc., USA).
Chlorophyll fluorescence changes are a useful index to measure photosynthetic efficiency. The fluorescence parameters were F 0 and F m—the initial or variable fluorescence yield, respectively, while the F v/F m ratio indicated a maximum potential of the plant’s photosynthetic ability. In our investigation, we used chlorophyll fluorometer OS-30P (Opti-Sciences, Inc., USA) according to Weng (2006). The data obtained were subjected to a statistical analysis and presented as mean ± SE. Student’s t test was used to calculate the significant differences as compared with the control.
Chloroplast detection using confocal microscopy
The collected tissue fragments were analyzed with using Nikon Eclipse TE300 confocal laser scanning inverted microscope. Obtained results were registered with an argon ion laser and He–neon laser emitting light with a wavelength of 488 nm (blue excitation and green fluorescence) and 543 nm (green excitation and red fluorescence), respectively. A mid pinhole, a long exposure time (90 s) and 100× (numerical aperture, 1.4) Plan Apochromat DCI H oil immersion lens were used in the analyses. We collected pairs of images in the green and red channels simultaneously. The EZ 2000 Viewer software package (Nikon Europe BV, Badhoevedorp, the Netherlands) was used for results documentation and analysis.
Determination of endogenous JAs
Endogenous jasmonates (jasmonic acid—JA, methyl jasmonate—JAMe) were determined with the method described by Wilmowicz et al. (2014) with modifications. Internal standards were 100 ng d2-JAMe and 100 ng d5-JA added to the crude extract. GC/MS-selected ion monitoring was performed by monitoring m/z 193, 195, 198, 224, 226 and 229.
Molecular cloning of InJMT cDNA
A full sequence of InJMT was obtained in PCR reactions using specific primers 5′-TTCCCCGAAAGCATGGGCA-3′ (forward); 5′-TCGGTTGTAGGATCGCCGGA-3′ (reverse) designed for EST fragments of InJMT (identified EST sequences of P. nil in the NCBI database: no. BJ560292.1 and CJ756546.1) (Supplementary Fig. 2). The reaction mixture was subjected to the following PCR conditions: 95 °C for 5 min, 1 cycle; 95 °C for 1 min, 62–50 °C for 30 s, 72 °C for 45 s for 15 cycles, followed by 1 cycle of incubation at 72 °C for 7 min. An amplified cDNA fragment was isolated from an agarose gel with the GeneMATRIX Agarose-Out DNA Purification Kit (EurX, Gdańsk, Poland) and sequenced by “Genomed S.A.” (Warsaw, Poland). The full-length InJMT sequence has been deposited in the GenBank database.
The bioinformatical analysis and phylogenetic tree calculation of identified cDNA was made with using ClustalW (http://www.ebi.ac.uk/clustalw), BLAST 2.2.25 (http://www.ncbi.nlm.nih.gov/BLAST) and ExPASy (http://www.expasy.org) and the CLC Main Workbench v.5.6.1 program.
Quantitative real-time PCR (qPCR)
The gene expression analysis was performed by Real-Time PCR (RT-qPCR) with a LightCycler 2.0 Carousel-Based System and LightCycler TaqMan Master Kit (ROCHE Diagnostics GmbH, Germany). The cDNA templates were prepared according to Frankowski et al. (2015), whereas gene-specific primers and UPL probes for the InJMT (GenBank acc. no. KF573520.1) were designed using Universal ProbeLibrary Assay Design Center (http://www.roche-applied-science.com/sis/rtpcr/upl) (Supplementary Table 1). InACT4 (GenBank acc. no. HM802138.2) was used as a reference gene. qPCR reaction was performed according to Frankowski et al. (2015). Three independent replicate RNA preparations were used for the analyses, and data are shown as mean ± SE of three samples for each stage.
Results
Darkness and JAMe induce cotyledon senescence in I. nil
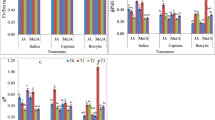
The progress of senescence was monitored by observing the inhibition of the functional activity of PSII (F v/F m), loss of chlorophyll and reduction of its autofluorescence. Our results showed that darkness promotes senescence through chlorophyll degradation (Fig. 1b) and autofluorescence (Fig. 2b), as well as a gradual decrease of the efficiency of PSII (Fig. 1a). The effect observed is accompanied by the progressive senescence processes. The F v/F m value in dark-grown plants and in light-grown plants treated with JAMe is 15 and 20 % lower than in control plants, respectively (Fig. 1a). The same solution applied to the cotyledons of plants grown under light conditions decreased the autofluorescence of the chlorophyll (Fig. 2c). JA has no influence on senescence in light-grown plants (Fig. 1a, b). Nevertheless, the application of MEF (a jasmonate biosynthesis inhibitor) on the cotyledons of I. nil reverses the stimulatory effect of the darkness on the process discussed. These plants contain a higher chlorophyll concentration than the cotyledons of plants grown in darkness and have a higher F v/F m ratio value (Fig. 1a, b), and also, the chlorophyll contained in them demonstrates autofluorescence similar to that of the control light-grown plants (Fig. 2d).
Changes in the maximum quantum efficiency of PSII [variable fluorescence (F v)/maximum fluorescence (F m) ratio] (a) and chlorophyll content (b) in the cotyledons of Ipomoea nil. CL continuous light, D darkness, JA jasmonic acid, JAMe jasmonic acid methyl ester, MEF mefenamic acid (a jasmonate biosynthesis inhibitor). Vertical bars indicate SE. Significant differences to the plants growing under light condition are indicated as **P < 0.01 and *P < 0.05
Visualization of chlorophyll autofluorescence in the cotyledons of Ipomoea nil growing under: continuous light (a), dark conditions (b), treated with jasmonic acid methyl ester after transfer to continuous light (c), treated with mefenamic acid (inhibitor of jasmonate biosynthesis) after transfer to darkness (d). Bars 200 µm
Isolation of the InJMT cDNA
The full-length InJMT cDNA (GB acc. no KF573520.1) obtained using the RT-PCR technique is composed of 1403 bp and encodes 380 amino acids (Fig. 3a, b). The predicted protein sequence of InJMT contains all the conserved domains characteristic of JMT identified in other plant species. A phylogenetic analysis of the amino acid sequence revealed that the predicted InJMT is very closely related to the JMT from Solanum tuberosum (StJMT, GB acc. no XP006341965.1) (Fig. 3c).
Identification of full-length InJMT cDNA. Products of PCR with specific primers (a). The coding sequence of InJMT and the deduced amino acid sequence (b). Subsequent nucleotide positions are marked on the left side of the figure, and amino acid positions on the right. The translation initiation point (the start codon) and termination point (the stop codon) are in boldface type. Small letters are used for regions not subject to translation. Domains responsible for methyl, salicylate, SAM (S-adenosyl methionine) and SAH (S-adenosyl-l-homocysteine) group binding are marked with light gray, dark gray, or are underlined, respectively. A phylogenetic analysis of InJMT with selected plant JMT sequences (c). Accession numbers of the sequences used to build the tree were as follows: RcJMT (XP002510424.1), SlJMT (XP004238275.1), StJMT (XP006341965.1), InJMT (AHA06001.1), VvJMT (XP002281588.1), TcJMT (XP007017600.1), PtJMT (XP002307671.1)
Senescence of I. nil cotyledons is accompanied by increase in the JAMe content and InJMT transcriptional activity
To investigate the possible role of JAs in cotyledon senescence in I. nil, we analyzed variations in the endogenous JA and JAMe level and the expression of InJMT coding for carboxyl methyltransferase during various stages of aging. The endogenous content of JAMe in the cotyledons of I. nil growing under darkness is initially hardly any lower than that of JA (Fig. 4b). However, the subsequent days see a gradual growth in the JAMe content in the cotyledons and a drop in the JA level. On day 8, the JAMe content is over four times that of JA, and over twice that of JAMe content in the control plants (Fig. 4b). The increased endogenous JAMe level is correlated with a stimulation of the InJMT transcriptional activity (Fig. 4a). Moreover, plants growing in darkness and treated with the jasmonate biosynthesis inhibitor (MEF) showed decrease in the InJMT expression and JAMe content (Fig. 4a, b).
The expression level of InJMT (related to InACT4) in the cotyledons of I. nil seedlings during darkness conditions and following MEF treatment (a). Dark-gray bars D conditions, gray bars MEF applied during darkness conditions. The endogenous jasmonates (JA jasmonic acid, JAMe jasmonic acid methyl ester) level in the cotyledons growing under darkness conditions and JAMe after MEF treatment (b). Mean ± SE n = 3. The thick horizontal line under the time axis indicates duration of the dark period. For JA, significant differences to the plants growing under SD condition are indicated as bb P < 0.01 and b P < 0.05. For JAMe, significant differences to the plants growing under SD condition are indicated as **P < 0.01 and *P < 0.05. For JAMe, after MEF treatment, significant differences to the plants growing under darkness condition are indicated as aa P < 0.01 and a P < 0.05
Discussion
Cotyledons are organs forming during embryogenesis as initially heterotrophic, after which they become photosynthetically active, and then, their function is to provide nutrients for seedlings. The senescence of cotyledons, like in the case of other organs, is an integral part of their development; similar to leaf senescence, it is genetically programmed and occurs when they already fulfilled their role. However, this process can be stimulated prematurely by stress factors, both biotic, e.g., pathogen infection, and abiotic, e.g., extreme temperatures, drought, nutrient deficiency, and shading (Krul 1974; Quirino et al. 2000). An important role in regulating this process is played by phytohormones, which are signal molecules and/or coordinators in the transformations accompanying senescence, such as nutrient recycling, the dismantling of chloroplasts, the cell death as a consequence of DNA, RNA and proteins degradation (Krul 1974; Peterman and Siedow 1985; Quirino et al. 2000; Wada et al. 2009). Among these phytohormones, jasmonates are particularly noteworthy. These substances cause chlorophyll loss, degradation of Rubisco and inhibition of its biosynthesis. They can also induce the disorganization of the thylakoid membrane system in chloroplasts, affect the functional activity of PSII, decrease the rates of transpiration and photosynthesis, as well as cell up-regulates several SAGs shown in different plants (Ananieva et al. 2004b; Beltrano et al. 1998; He and Gan 2001; He et al. 2002; Liu et al. 2015; Weaver et al. 1998; Weidhase et al. 1987a, b; Woo et al. 2001; Oh et al. 1996). Jasmonates have been widely shown to be powerful promoters of leaf senescence (Chen and Kao 1998; Chou and Kao 1992; Hung and Kao 1996; Tsai et al. 1996; Ueda and Kato 1981; Weidhase et al. 1987a), but their participation in cotyledon senescence is still poorly understood (Ananieva et al. 2004a, 2007). In the present study, we evaluate the effects of darkness and JAs on the senescence of I. nil cotyledons.
We showed that darkness causes progressive reduction in the content of chlorophyll in the cotyledons (Fig. 1b) and decreases its autofluorescence (Fig. 2c), diminishing the F v/F m ration, as well (Fig. 1a). In many plant species, the stimulating effect of darkness on senescence processes was observed (Biswal et al. 1983; Biswal and Biswal 1984; Oh et al. 1996; Weaver et al. 1998). Reducing the endogenous jasmonate content by applying the jasmonate biosynthesis inhibitor reverses dark-stimulated cotyledon senescence in I. nil (Figs. 1a, b, 2d), which confirms that these phytohormones play a key role in regulating this process. Although the conjugate of JA and the amino acid isoleucine (Ile) has been reported to be a bioactive form of jasmonates, the results of substantial physiological and molecular research do not eliminate a specific role played by other jasmonates, such as JAMe, in controlling of growth and developmental processes (Noir et al. 2013; Pak et al. 2009; Staswick 1992). Moreover, JAMe is a plant volatile regulator and is thought to be a potential candidate for intra- and intercellular signal transducers mediating jasmonate-dependent plant responses because of its ability to diffuse through the membranes (Seo et al. 2001; Cheong and Choi 2003).
JAMe applied to the cotyledons of light-grown I. nil accelerates their senescence. Both the chlorophyll content (Fig. 1b) and the F v/F m ratio (Fig. 1a) assume values lower than in plants with dark-stimulated senescence. No such effect was observed following JA applications (Fig. 1a, b). JAs, i.e., both JA and JAMe, are promoters of leaf senescence upon their exogenous application (Creelman and Mullet 1997; Wasternack and Hause 2002). JAMe is the most effective stimulator of leaf senescence in the leaves of wild-type and mutant Arabidopsis (Oh et al. 1996; Weaver et al. 1998; Woo et al. 2001), leaf segments of barley (Reinbothe et al. 1993, 1997; Weidhase et al. 1987a) and intact cotyledons of Cucurbita pepo (zucchini) (Ananieva et al. 2004b).
Darkness leads to increased concentrations of endogenous JAMe and decreases the pool of free JA (Fig. 4b), which shows that JAMe is a cotyledon senescence stimulator in I. nil. If a change in the JAMe level is important for explaining cotyledon senescence, it is crucial to study the expression level of the InJMT gene that encodes a cytosolic protein involved in the methylation of JA. In the present investigation, we identified this gene in I. nil (Fig. 3). Based on a comparison between the deduced amino acid sequences of InJMT and JMTs from other plant species, it was found that InJMT contains all of the characteristic motifs present in SAM and SAM-like methyltransferases, SAM/SAH binding residues, salicylate/JA binding residues and additional active site residues (Fig. 3b, c), which are essential for its activity (Martin and McMillan 2002). Bearing this in mind, it can be suspected that the gene encodes a functional enzymatic protein. A progressing senescence of I. nil cotyledons is correlated with a growth in InJMT expression (Fig. 4a), which elevates the JAMe content (Fig. 4b). The drop in the endogenous JAMe content on day 8 of darkness may be caused by the exhaustion of the available substrate for the enzymatic activity of InJMT.
These results suggest that the accelerated cotyledon senescence in dark-grown I. nil is correlated with an increased InJMT expression level and, consequently, a growth in the concentration of JAMe which plays the role of a signal molecule in regulating this process.
Author contribution statement
Emilia Wilmowicz and Agata Kućko designed and carried out all the experiments and wrote the manuscript. Kamil Frankowski helped with gene identification and qPCR reactions, Michał Świdziński conducted the microscopy experiments, Katarzyna Marciniak measured the chlorophyll content and quantum efficiency of PS II, Jan Kopcewicz and Agata Kućko were responsible for preparing the final version of manuscript. All authors were responsible for analysis and interpretation of obtained results.
References
Acosta IF, Farmer EE (2010) Jasmonates. Arabidopsis Book 8:e0129
Ananieva K, Malbeck J, Kamínek M, Van Staden J (2004a) Methyl jasmonate down-regulates endogenous cytokinin levels in cotyledons of Cucurbita pepo (zucchini) seedlings. Physiol Plant 122:496–503
Ananieva K, Malbeck J, Kamínek M, Van Staden J (2004b) Changes in endogenous cytokinin levels in cotyledons of Cucurbita pepo (zucchini) during natural and dark induced senescence. Physiol Plant 122:133–142
Ananieva K, Ananieva ED, Mishev K, Georgieva K, Malbeck J, Kamínek M, Van Staden J (2007) Methyl jasmonate is a more effective senescence-promoting factor in Cucurbita pepo (zucchini) cotyledons when compared with darkness at the early stage of senescence. J Plant Physiol 164:1179–1187
Ananieva K, Ananieva ED, Doncheva S, Georgieva K, Tzvetkova N, Kamínek M, Motyka V, Dobrev P, Gajdosová S, Malbeck J (2008a) Senescence progression in a single darkened cotyledon depends on the light status of the other cotyledon in Cucurbita pepo (zucchini) seedlings: potential involvement of cytokinins and cytokinin oxidase/dehydrogenase activity. Physiol Plant 134:609–623
Ananieva K, Ananieva ED, Mishev K, Georgieva K, Tzvetkova N, Van Staden J (2008b) Changes in photosynthetic capacity and polypeptide patterns during natural senescence and rejuvenation of Cucurbita pepo L. (zucchini) cotyledons. Plant Growth Regul 54:23–29
Avice JC, Etienne P (2014) Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.). J Exp Bot 65:3813–3824
Beltrano J, Ronco MG, Montaldi ER, Carbone A (1998) Senescence of flag leaves and ears of wheat hastened by methyl jasmonate. J Plant Growth Regul 17:53–57
Biswal UC, Biswal B (1984) Photocontrol of leaf senescence. Photochem Photobiol 39:875–879
Biswal B, Choudhury NK, Sahu P, Biswal UC (1983) Senescence of detached fern leaves. Plant Cell Physiol 24:1203–1208
Bouchard JN, Yamasaki H (2008) Heat stress stimulates nitric oxide production in Symbiodinium microadriaticum: a possible linkage between nitric oxide and the coral bleaching phenomenon. Plant Cell Physiol 49:641–652
Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, Zhang C, Morris K, Jenner C, Jackson S, Thomas B, Tabrett A, Legaie R, Moore JD, Wild DL, Ott S, Rand D, Beynon J, Denby K, Mead A, Buchanan-Wollaston V (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23:873–894
Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42:567–585
Chen SJ, Kao CH (1998) Methyl jasmonate, ammonium, and leaf senescence in rice. J Plant Physiol 152:353–357
Cheong JJ, Choi YD (2003) Methyl jasmonate as a vital substance in plants. Trends Genet 19:409–413
Chou CM, Kao CH (1992) Methyl jasmonate, calcium, and leaf senescence in rice. Plant Physiol 99:1693–1694
Corpas FJ, Hayashi M, Mano S, Nishimura M, Barroso JB (2009) Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiol 151:2083–2094
Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48:355–381
Du J, Li M, Kong D, Wang L, Lv Q, Wang J, Bao F, Gong Q, Xia J, He Y (2014) Nitric oxide induces cotyledon senescence involving co-operation of the NES1/MAD1 and EIN2-associated ORE1 signalling pathways in Arabidopsis. J Exp Bot 65:4051–4063
Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132:4563–4574
Frankowski K, Kęsy J, Wojciechowski W, Kopcewicz J (2009) Light- and IAA-regulated ACC synthase gene (PnACS) from Pharbitis nil and its possible role in IAA-mediated flower inhibition. J Plant Physiol 166:192–202
Frankowski K, Wilmowicz E, Kućko A, Zienkiewicz A, Zienkiewicz K, Kopcewicz J (2015) Profiling the BLADE-ON-PETIOLE gene expression in the abscission zone of generative organs in Lupinus luteus. Acta Physiol Plant 37:220. doi:10.1007/s11738-015-1972-y
Gan S (2004) The hormonal regulation of senescence. In: Davies PJ (ed) Plant hormones: biosynthesis, signal transduction and action. Kluwer Academic Publishers, Dordrecht, pp 561–581
Glazińska P, Wilmowicz E, Wojciechowski W, Frankowski K, Kopcewicz J (2014) Impact of InMIR319 and light on the expression of InTCP4 gene involved in the development of Ipomoea nil plants. Acta Physiol Plant 36:29–43
Gombert J, Etienne P, Ourry A, Le Dily F (2006) The expression patterns of SAG12/Cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. J Exp Bot 57:1949–1956
He Y, Gan S (2001) Identical promoter elements are involved in regulation of the OPR1 gene by senescence and jasmonic acid in Arabidopsis. Plant Mol Biol 47:595–603
He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128:876–884
Hung KT, Kao CH (1996) Promotive effect of jasmonates on the senescence of detached maize leaves. Plant Growth Regul 19:77–83
Hung KT, Kao CH (2004) Hydrogen peroxide is necessary for abscisic acid-induced senescence of rice leaves. J Plant Physiol 161:1347–1357
Jing HC, Schippers JH, Hille J, Dijkwel PP (2008) Ethylene induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J Exp Bot 56:2915–2923
Kanazawa S, Sano S, Koshiba T, Ushimaru T (2000) Changes in antioxidative enzymes in cucumber cotyledons during natural senescence: comparison with those during dark-induced senescence. Physiol Plant 109:211–216
Kęsy J, Wilmowicz E, Maciejewska B, Frankowski K, Glazińska P, Kopcewicz J (2011) Independent effects of jasmonates and ethylene on inhibition of Pharbitis nil flowering. Acta Physiol Plant 33:1211–1216
Krul WR (1974) Nucleic acid and protein metabolism of senescing and regenerating soybean cotyledons. Plant Physiol 54:36–40
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136
Lin M, Pang C, Fan S, Song M, Wei H, Yu S (2015) Global analysis of the Gossypium hirsutum L. Transcriptome during leaf senescence by RNA-Seq. BMC Plant Biol 12(15):43. doi:10.1186/s12870-015-0433-5
Liu L, Li H, Zeng H, Cai Q, Zhou X, Yin Ch (2015) Exogenous jasmonic acid and cytokinin antagonistically regulate rice flag leaf senescence by mediating chlorophyll degradation, membrane deterioration, and senescence-associated genes expression. J Plant Growth Regul. doi:10.1007/s00344-015-9539-0
Lu C, Koroleva OA, Farrar JF, Gallagher J, Pollock CJ, Deri TA (2002) Rubisco small subunit, chlorophyll a/b-binding protein and sucrose:fructan-6-fructosyl transferase gene expression and sugar status in single barley leaf cells in situ. Cell type specificity and induction by light. Plant Physiol 130:1335–1348
Ma W, Smigel A, Tsai YC, Braam J, Berkowitz GA (2008) Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol 148:818–828
Maciejewska B, Kopcewicz J (2002) Inhibitory effect of methyl jasmonate on flowering and elongation growth in Pharbitis nil. J Plant Growth Regul 21:216–223
Maciejewska BD, Kęsy J, Zielińska M, Kopcewicz J (2004) Jasmonates inhibit flowering in short-day plant Pharbitis nil. Plant Growth Regul 43:1–8
Martin JL, McMillan FM (2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol 12:783–793
McKersie BD, Lepock JR, Kruuv J, Thompson JE (1987) The effects of cotyledon senescence on the composition and physical properties of membrane lipid. Biochim Biophys Acta 508:197–212
Neill S, Barros R, Bright J, Desikan R, Hancock J, Harrison J, Morris P, Ribeiro D, Wilson I (2008) Nitric oxide, stomatal closure, and abiotic stress. J Exp Bot 59:165–176
Noir S, Bömer M, Takahashi N, Ishida T, Tsui TL, Balbi V, Shanahan H, Sugimoto K, Devoto A (2013) Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol 161:1930–1951
Norastehnia A, Sajedi RH, Nojavan-Asghari M (2007) Inhibitory effects of methyl jasmonate on seed germination in maize (Zea mays): effect on α-amylase activity and ethylene production. Gen Appl Plant Physiol 33:13–23
Oh SA, Lee SY, Chung IK, Lee CH, Nam HG (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30:739–754
Okushima Y, Mitina I, Quach HL, Theologis A (2005) AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J 43:29–46
Pak H, Guo Y, Chen M, Chen K, Li Y, Hua S, Shamsi I, Meng H, Shi C, Jiang L (2009) The effect of exogenous methyl jasmonate on the flowering time, floral organ morphology, and transcript levels of a group of genes implicated in the development of oilseed rape flowers (Brassica napus L.). Planta 231:79–91
Peterman TK, Siedow JN (1985) Behavior of lipoxygenase during establishment, senescence, and rejuvenation of soybean cotyledons. Plant Physiol 78:690–695
Pružinská A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania KH, Kräutler B, Youn JY, Liljegren SJ, Hörtensteiner S (2005) Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol 139:52–56
Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5:278–282
Reinbothe S, Reinbothe C, Parthier B (1993) Methyl jasmonate represses translation initiation of a specific set of mRNAs in barley. Plant J 4:459–467
Reinbothe C, Parthier B, Reinbothe S (1997) Temporal pattern of jasmonate-induced alterations in gene expression of barley leaves. Planta 201:281–287
Rukes KL, Mulkey TJ (1993) Hormonal regulation of cotyledon senescence. Bioscience 19:10–15
Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98:4788–4793
Staswick P (1992) Jasmonate, genes and fragrant signals. Plant Physiol 99:804–807
Troncoso-Poncea MA, Caoc X, Yangb Z, Ohlrogge JB (2013) Lipid turnover during senescence. Plant Sci 205–206:13–19
Tsai FY, Hung KT, Kao CH (1996) An increase in ethylene sensitivity is associated with jasmonate-promoted senescence of detached rice leaves. J Plant Growth Regul 15:197–200
Ueda J, Kato J (1981) Promotive effect of methyl jasmonate on oat leaf senescence in the light. Z Pflanzenphysiol 103:357–359
Van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141:776–792
Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149:885–893
Wang CY (1998) Methyl jasmonate inhibits postharvest sprouting and improves storage quality of radishes. Postharvest Biol Technol 14:179–183
Wasternack C, Hause B (2002) Jasmonates and octadecanoids: signals in plant stress responses and plant development. Prog Nucleic Acid Res Mol Biol 72:165–221
Wasternack C, Kombrink E (2010) Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem Biol 5:63–77
Watanabe A, Hamada K, Yokoi H, Watanabe A (1994) Biphasic and differential expression of cytosolic glutamine synthetase genes of radish during seed germination and senescence of cotyledons. Plant Mol Biol 26:1807–1817
Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37:455–469
Weidhase RA, Kramell HM, Lehmann J, Liebisch HW, Lerbs W, Parthier B (1987a) Methyl jasmonate-induced changes in the polypeptide pattern of senescing barley leaf segments. Plant Sci 51:177–186
Weidhase RA, Lehmann J, Kramell H, Sembdner G, Parthier B (1987b) Degradation of ribulose-15-bisphosphate-carboxylase and chlorophyll in senescing barley leaf segments triggered by jasmonic acid methyl ester and counteraction by cytokinin. Physiol Plant 69:161–166
Weng JH (2006) Underestimate of PS2 efficiency in the field due to high leaf temperature resulting from leaf clipping and its amendment. Photosynthetica 44:467–470
Wilmowicz E, Kęsy J, Kopcewicz J (2008) Ethylene and ABA interactions in the regulation of flower induction in Pharbitis nil. J Plant Physiol 165:1917–1928
Wilmowicz E, Frankowski K, Grzegorzewska W, Kęsy J, Kućko A, Banach M, Szmidt-Jaworska A, Saniewski M (2014) The role of jasmonates in the formation of a compound of chalcones and flavans with phytoalexin-like properties in mechanically wounded scales of Hippeastrum × hybr. bulbs. Acta Biol Crac Ser Bot 56:54–58
Wilson ZA, Song J, Taylor B, Yang C (2011) The final split: the regulation of anther dehiscence. J Exp Bot 62:1633–1649
Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13:1779–1790
Wrischer M, Prebeg T, Magnus V, Ljubešić N (2009) Unusual thylakoid structures appearing during degradation of the photosynthetic apparatus in chloroplasts. Acta Bot Croat 68:1–9
Xuan Y, Zhou S, Wang L, Cheng Y, Zhao L (2010) Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiol 153:1895–1906
Zhang MP, Zhang CJ, Yu GH, Jiang YZ, Strasser RJ, Yuan ZY, Yang XS, Chen GX (2010) Changes in chloroplast ultrastructure, fatty acid components of thylakoid membrane and chlorophyll a fluorescence transient in flag leaves of a super-high-yield hybrid rice and its parents during the reproductive stage. J Plant Physiol 167:277–285
Zhao MG, Chen L, Zhang LL, Zhang WH (2009) Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol 151:755–767
Acknowledgments
This project was supported by Nicolaus Copernicus University Grant No. 1233-B and funds provided by Nicolaus Copernicus University (Toruń, Poland) for the research program of the Chair of Plant Physiology and Biotechnology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. K. Nagar.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wilmowicz, E., Kućko, A., Frankowski, K. et al. Methyl jasmonate-dependent senescence of cotyledons in Ipomoea nil . Acta Physiol Plant 38, 222 (2016). https://doi.org/10.1007/s11738-016-2244-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-016-2244-1