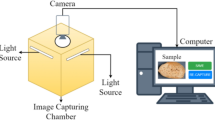
Abstract
Melon fruit phenotype has rich genetic variation and is an important target trait for breeding and commerciality. Existing measurement techniques have problems such as low detection accuracy, low efficiency and vague quantification methods. The study uses a deep learning approach to build an improved U-Net network for accurate segmentation of melon pericarp and seed cavity characteristics, as well as quantitative computation of phenotypic characteristics. The improved network uses VGG16 to replace the encoder in the original U-Net network, migrates the pre-training weights under the PASCAL VOC dataset, and adds a Repeated Criss-Cross Attention to the U-Net network skip connection to improve the network training efficiency and enhance the acquisition of image contextual information to solve the problem of few melon samples and poor fruit skin cavity segmentation. The segmentation results were further quantified to obtain precise parameters of melon fruit phenotypic characteristics. The experiments showed that the segmentation accuracy indexes of the improved U-Net network were 98.83% for Mean Pixel Accuracy and 96.67% for Mean Intersection over Union, which were higher than those of other comparative algorithms. The relative error of each quantitative measurement parameter is less than 5%, and the repeat accuracy is less than 0.1%, which has high accuracy and robustness. It meets the requirements of rapid detection of melon fruit characteristics and provides a powerful help for the accurate acquisition of its germplasm resource information.









Similar content being viewed by others
References
G. Zhao, Q. Lian, Z. Zhang et al., A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 51, 1607–1615 (2019). https://doi.org/10.1038/s41588-019-0522-8
P. Gao, S. Liu, H.N. Cui, T.F. Zhang, X.Z. Wang, H.Y. Liu, Z.C. Zhu, F.S. Luan, Research progress of melon genomics, functional gene mapping and genetic engineering. Acta Hortic. Sin. 47(09), 1827–1844 (2020). https://doi.org/10.16420/j.issn.0513-353x.2020-0489
J.B. Hu, S.W. Ma, J.M. Wang, Y. Su, Q. Li, Establishment of a melon (Cucumis melo) core collection based on phenotypic characters. J. Fruit Sci. 30(03), 404–411 (2013). https://doi.org/10.13925/j.cnki.gsxb.2013.03.015
H.F. Sun, Application of breeding technology in modern agricultural production. Grain Sci. Technol. Econ. 44(12), 77–80 (2019). https://doi.org/10.16465/j.gste.cn431252ts.20191221
G. Qiu, L. Liu, X.M. Li, X.Z. Wang, Research progress on breeding for resistance to fusarium wilt and powdery mildew in muskmelon. Biotechnol. Bull. 33(08), 14–19 (2017). https://doi.org/10.13560/j.cnki.biotech.bull.1985.2017-0470
W.K. Jia, H. Meng, X.H. Ma, Y.N. Zhao, Z. Ji, Y.J. Zhng, Efficient detection model of green target fruit based on optimized transformer network. Trans. Chin. Soc. Agric. Eng. 37(14), 163–170 (2021)
J.L. Chen, X.P. Deng, S.H. Nie, Development status and strategic analysis of smart agriculture. Agric. Eng. Technol. 40(33), 64+69 (2020). https://doi.org/10.16815/j.cnki.11-5436/s.2020.33.020
R.A. Arun, S. Umamaheswari, A.V. Jain, Reduced U-Net architecture for classifying crop and weed using pixel-wise segmentation, in INOCON (2020), pp. 1–6. https://doi.org/10.1109/INOCON50539.2020.9298209
Y. Ai, C. Sun, J. Tie, J. Tie, Research on recognition model of crop diseases and insect pests based on deep learning in harsh environments. IEEE Access 8, 171686–171693 (2020). https://doi.org/10.1109/ACCESS.2020.3025325
X.K. Dai, X.S. Wang, L.H. Du, N. Ma, S.P. Xu, B.N. Cai, S.X. Wang, Z.G. Wang, B.L. Qu, Automatic segmentation of head and neck organs at risk based on three-dimensional U-Net deep convolutional neural network. J. Biomed. Eng. 37(01), 136–141 (2020)
H.W. Mou, Y. Guo, X.H. Quan, Z.M. Cao, J. Han, Magnetic resonance imaging brain tumor image segmentation based on improved U-Net. Laser Optoelectron. Prog. 58(04), 265–272 (2021)
D. Yang, G.R. Liu, M.C. Ren, H.Y. Pei, Retinal blood vessel segmentation method based on multi-scale convolution kernel U-Net model. J. Northeast. Univ. (Nat. Sci.) 42(01), 7–14 (2021)
S. Karen, Z. Andrew, Very deep convolutional networks for large-scale image recognition. CoRR. (2015). https://arxiv.org/abs/1409.1556
E. Shelhamer, J. Long, T. Darrell, Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(04), 640–651 (2017). https://doi.org/10.1109/TPAMI.2016.2572683
H. Zhao, J. Shi, X. Qi, X. Wang, J. Jia, Pyramid scene parsing network, in CVPR (2017), pp. 6230–6239. https://doi.org/10.1109/CVPR.2017.660
L.C. Chen, G. Papandreou, F. Schroff, H. Adam, Rethinking atrous convolution for semantic image segmentation. Computer Vision and Pattern Recognition (2017). https://arxiv.org/abs/1706.05587
Z. Huang, X. Wang, Y. Wei, L. Huang, H. Shi, W. Liu, T.S. Huang, CCNet: criss-cross attention for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. (2020). https://doi.org/10.1109/TPAMI.2020.3007032
O. Ronneberger, P. Fischer, T. Brox, U-Net: convolutional networks for biomedical image segmentation. Med. Image Comput. Comput. Assist. Interv. 9351, 234–241 (2015). https://doi.org/10.1007/978-3-319-24574-4\_28
Q. Pan, M. Gao, P. Wu, J. Yan, S. Li, A deep-learning-based approach for wheat yellow rust disease recognition from unmanned aerial vehicle images. Sensors (2021). https://doi.org/10.3390/s21196540
N. Stasenko, E. Chernova, D. Shadrin, I. Ovchinnikov, M. Pukalchik, Deep learning for improving the storage process: accurate and automatic segmentation of spoiled areas on apples, in 2021 IEEE International Instrumentation and Measurement Technology Conference (I2MTC) (2021). https://doi.org/10.1109/I2MTC50364.2021.9460071
J. Chen, M. Han, Y. Lian, S. Zhang, Segmentation of impurity rice grain images based on U-Net model. Trans. Chin. Soc. Agric. Eng. 36(10), 174–180 (2020)
Z.Q. Fang, H.G. Xiong, S.H. Xiao, G.F. Li, Regular workpiece measurement system with multiple plane dimensions based on monocular vision. Mach. Des. Manuf. 11, 241-245+249 (2020). https://doi.org/10.19356/j.cnki.1001-3997.2020.11.060
J. Weng, P. Cohen, M. Herniou, Camera calibration with distortion models and accuracy evaluation. IEEE Trans. Pattern Anal. Mach. Intell. 14(10), 965–980 (1992). https://doi.org/10.1109/34.159901
Z. Zhang, A flexible new technique for camera calibration. IEEE Trans. Pattern Anal. Mach. Intell. 22(11), 1330–1334 (2000). https://doi.org/10.1109/34.888718
J. Kang, J.M. Ding, Y. Wang, T. Lei, Liver image segmentation algorithm based on watershed correction and U-Net. Comput. Eng. 46(01), 255-261+270 (2020). https://doi.org/10.19678/j.issn.1000-3428.0055495
N.B. Yu, J.N. Liu, L. Gao, Z.W. Sun, J.D. Han, Auto-segmentation method based on deep learning for the knee joint in MR images. Chin. J. Sci. Instrum. 41(06), 140–149 (2020). https://doi.org/10.19650/j.cnki.cjsi.J2006199
R.R. Zhang, L. Xia, L.P. Chen, C.C. Xie, M.X. Chen, W.J. Wang, Recognition of wilt wood caused by pine wilt nematode based on U-Net network and unmanned aerial vehicle images. Trans. Chin. Soc. Agric. Eng. 36(12), 61–68 (2020)
M. Ye, N. Ruiwen, Z. Chang, G. He, H. Tianli, L. Shijun, S. Yu, Z. Tong, G. Ying, A lightweight model of VGG-16 for remote sensing image classification. IEEE J Sel. Top. Appl. Earth Obs. Remote Sens. 14, 6916–6922 (2021). https://doi.org/10.1109/JSTARS.2021.3090085
T. Lee, L. Huang, P. Kuo, C. Wang, J. Guo, Focal-balanced attention U-Net with dynamic thresholding by spatial regression for segmentation of aortic dissection in CT imagery, in 2021 IEEE 18th International Symposium on Biomedical Imaging (2021), pp. 541–544. https://doi.org/10.1109/ISBI48211.2021.9434028
T. Falk, D. Mai, R. Bensch et al., U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019). https://doi.org/10.1038/s41592-018-0261-2
L.K. Li, Z.H. Lu, B. Zou, Research on target detection and feasible region segmentation based on deep learning. Laser Optoelectron. Prog. 57(12), 189–197 (2020)
R. Renaguli, S.Z. Liu, T.J. Liu, B.Z. Chen, J.S. Wang, Survey of plant leaf area measurement methods. Anhui Agric. Sci. Bull. 26(05), 22–23 (2020). https://doi.org/10.16377/j.cnki.issn1007-7731.2020.05.009
Acknowledgements
This work was supported by Key Laboratory of Storage of Agricultural Products, Ministry of Agriculture and Rural Affairs [Project Number: Kf2021003]; Natural Science Foundation of Hebei Province [Project Number: 2021202136]; Tianjin Science and Technology Plan Project [Project Number: 21ZYCGSN00320].
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qian, C., Liu, H., Du, T. et al. An improved U-Net network-based quantitative analysis of melon fruit phenotypic characteristics. Food Measure 16, 4198–4207 (2022). https://doi.org/10.1007/s11694-022-01519-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11694-022-01519-7