Abstract
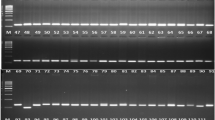
We analyzed the genetic differences of 16 poplar clones between genomic-SSR and EST-SSR markers. The statistical results show that the average number of alleles detected by genomic-SSR was 4.1, Shannon’s index 1.0646, observed heterozygosity 0.4427 and expected heterozygosity 0.5523, while for the EST-SSR, the average number of alleles was 2.8, Shannon’s index 0.6985, observed heterozygosity 0.2330 and expected heterozygosity 0.4684. Cluster analysis indicated that the EST-SSR capacity of genotypic identification was more precise than that of genomic-SSR. These results reveal that EST-SSR and genomic-SSR have statistically significant genetic differences in polymorphism detection and genotypic identification. These differences could provide a theoretical basis for the rational use of SSR markers in species diversity and other related research.
Similar content being viewed by others
References
Ayres N M, McClung A M, Larkin P D, Bligh H F J, Jones C A, Park W D. 1997. Microsatellites and a single-nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germ plasm. Theor Appl Genet, 94: 773–781
Botstein D, White R L, Skolnick R M, Davis R W. 1980. Construction of genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet, 32: 314–331
Chang W, Zhao X, Li X, Qiu B, Han Y P, Teng W L, Li W B. 2009. Development of soybean EST-SSR marker and comparison with genomic-SSR marker. Chin J Oil Crop Sci, 31(2): 149–156 (in Chinese with English abstract)
Dellaporta S L, Wood J, Hicks J. 1983. A rapid method for DNA extraction from plant tissue. Plant Mol Biol Rep, 1: 19–21
Ellis J R, Burke J M. 2007. EST-SSRs as a resource for population genetic analyses. Heredity, 99: 125–132
Eujayl I, Sorrells M, Baum M, Wolters P, Powell W. 2001. Assessment of genotypic variation among cultivated durum wheat based on EST-SSRs and genomic-SSRs. Euphytica, 119: 39–43
Hanai L R, Santini L, Camargo L E A, Fungaro M H P, Gepts P, Tsai S M, Vieira M L C. 2010. Extension of the core map of common bean with EST-SSR, RGA, AFLP, and putative functional markers. Mol Breeding, 25: 25–45
Jia J Z, Zhang Z B, Devos K, Gale M D. 2001. Analysis of genetic diversity of 21 chromosomes of wheat by RFLP mapping lotus. Sci Chin (Ser C), 31(1): 13–21 (in Chinese)
Maynard Smith J, Haigh J. 1974. The hitch-hiking effect of a favorable gene. Genet Res, 23(1): 23–35
Moccia M D, Oger-Desfeux C, Marais G A B, Widmer A. 2009. A White Campion (Silene latifolia) floral expressed sequence tag (EST) library: annotation, EST-SSR characterization, transferability, and utility for comparative mapping. BMC Genom, 10: 243–257
Nei M, Li W H. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA, 76: 5269–5273
Su X H, Huang Q J, Zhang B Y, Zhang X H. 2004. The achievement and developing strategy on variety selection and breeding of poplar in China. World Forest Res, 17(1): 46–49 (in Chinese with English abstract)
Sun Q X, Huang T C, Ni Z F, Procunier J D. 1996. Study on wheat heterotic group I. Genetic diversity revealed by random amplified polymorphic DNA (RAPD) in elite wheat cultivars. J Agric Biotechnol, 4(2): 103–110 (in Chinese with English abstract)
Tuskan G A, Gunter L E, Yang Z K, Yin T M, Sewell M M, Di-Fazio S P. 2004. Characterization of microsatellites revealed by genomic sequencing of Populus trichocarpa. Can J Forest Res, 34: 85–93
Varshney R K, Graner A, Sorrells M E. 2005. Genic microsatellite markers in plants: features and applications. Trends Biotechnol, 23(1): 48–55
Wen M F, Wang H Y, Xia Z Q, Zou M L, Lu C, Wang W Q. 2010. Development of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha curcas L. BMC Res Notes, 3: 42–50
Yang X Q, Liu P, Han Z F, Ni Z F, Liu W Q, Sun Q X. 2005. Comparative analysis of genetic diversity revealed by genomic-SSR, EST-SSR and pedigree in wheat (Triticum asetivum L.). Acta Genet Sin, 32(4): 406–416 (in Chinese with English abstract)
Yang Y L, Zhang Y D, Zhang X Y. 2008. Transferability analysis of Populus SSR markers in Salix. Mol Plant Breeding, 6(6): 1134–1138 (in Chinese with English abstract)
Yin T M, DiFazio S P, Gunter L E, Zhang X Y, Sewell M M, Woolbright S A, Allan G J, Kelleher C T, Douglas C J, Wang M X, Tuskan G A. 2008. Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res, 18: 422–430
Zeng S H, Xiao G, Guo J, Fei Z J, Xu Y Q, Roe B A, Wang Y. 2010. Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et Zucc.) Maxim. BMC Genom, 11: 94–105
Zhao Y, Kong F M, Ding C Q, Li S S. 2008. Transferability of EST-SSR markers in zoysiagrasses. Chin J Grassland, 30(3): 69–73 (in Chinese with English abstract)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, Yp., Jiang, Xb., Zhang, M. et al. Differences of EST-SSR and genomic-SSR markers in assessing genetic diversity in poplar. For. Stud. China 14, 1–7 (2012). https://doi.org/10.1007/s11632-012-0106-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11632-012-0106-5