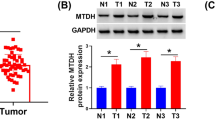
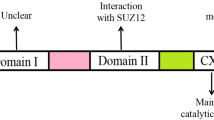
Summary
Histone deacetylase was overexpressed in a variety of cancers and was closely correlated with oncogenic factors. The histone deacetylase inhibitor, trichostatin A (TSA) was shown to induce apoptosis in many cancer cells. However, the mechanism of TSA on induction of cancer cells apoptosis is poorly understood. This study was designed to characterize the global gene expression profiles before and after treatment of human leukemia cell line Molt-4 with TSA. Flow cytometry, MTT and DNA ladder were used to observe the effect of TSA on the apoptosis of MOLT-4 cells and normal human peripheral blood mononuclear cells (PBMC). Microarray, reverse transcription-polymerase chain reaction (RT-PCR) and Western blotting were used to detect the difference of gene and protein expressions of Molt-4 cells after incubation of the cells with TSA. The results showed that TSA could induce Molt-4 apoptosis in dose- and time-dependent manners but spared PBMCs. Microarray analysis showed that after incubation with TSA for 9 h, 310 genes were upregulated and 313 genes were deregulated. These genes regulate the growth, differentiation and survival of cells. Among these genes, STAT5A was down-regulated by 80.4% and MYC was down-regulated by 77.3%. It was concluded that TSA has definite growth-inhibiting and apoptosis-inducing effects on Molt-4 cells in time- and dose-dependent manners, with weak cytotoxic effects on PBMCs at the same time. The mechanism of TSA selectively inducing apoptosis and inhibiting growth may be ascribed to the changes of pro-proliferation genes and anti-apoptosis genes.
Similar content being viewed by others
References
Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer, 2006,6(1):38–51
De los Santos M, Zambrano A, Aranda A. Combined effects of retinoic acid and histone deacetylase inhibitors on human neuroblastoma SH-SY5Y cells. Mol Cancer Ther, 2007,6(4):1425–1432
Vlasáková J, Nováková Z, Rossmeislová L, et al. Histone deacetylase inhibitors suppress IFNalpha-induced up-regulation of promyelocytic leukemia protein. Blood, 2007,109(4):1373–1380
Dalgard CL, Van Quill KR, O’Brien JM. Evaluation of the in vitro and in vivo antitumor activity of histone deacetylase inhibitors for the therapy of retinoblastoma. Clin Cancer Res, 2008,14(10):3113–3123
Noh EJ, Lim DS, Jeong G, et al. An HDAC inhibitor, trichostatin A, induces a delay at G2/M transition, slippage of spindle checkpoint, and cell death in a transcription-dependent manner. Biochem Biophys Res Commun, 2009,378(3):326–331
Chen WK, Chen Y, Gu JX, et al. Effect of trichostatin A on histone acetylation level and apoptosis in HL-60 cells. J Exp Hematol, 2004,12(3):324–328
Zhou J, Gao Q, Chen G, et al. Novel oncolytic adenovirus selectively targets tumor-associated polo-like kinase 1 and tumor cell viability. Clin Cancer Res, 2005,11(23): 8431–8440
Miyanaga A, Gemma A, Noro R, et al. Antitumor activity of histone deacetylase inhibitors in non-small cell lung cancer cells: development of a molecular predictive model. Mol Cancer Ther, 2008,7(7):1923–1930
Roy S, Shor AC, Bagui TK, et al. Histone deacetylase 5 represses the transcription of cyclin D3. J Cell Biochem, 2008,104(6):2143–2154
Yu H, Jove R. The STATs of cancer-new molecular targets come of age. Nat Rev Cancer, 2004,4(2):97–105
Ihle JN. STATs: signal transducers and activators of transcription. Cell, 1996,84:331–334
Darnell JE Jr. STATs and gene regulation. Science, 1997, 277:1630–1635
Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci, 2004,117(Pt 8):1281–1283
Defilippi P, Rosso A, Dentelli P, et al. {beta} 1 Integrin and IL-3R coordinately regulate STAT5 activation and anchorage-dependent proliferation. J Cell Biol, 2005,168(7):1099–1108
Sebastián C, Serra M, Yeramian A, et al. Deacetylase activity is required for STAT5-dependent GM-CSF functional activity in macrophages and differentiation to dendritic cells. J Immunol, 2008,180(9):5898–5906
Wierenga AT, Vellenga E, Schuringa JJ. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol Cell Biol, 2008,28(21): 6668–6680
Harir N, Boudot C, Friedbichler K, et al. Oncogenic kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood, 2008,112(6): 2463–2473
Zeoli A, Dentelli P, Rosso A, et al. Interleukin-3 promotes expansion of hemopoietic-derived CD45+ angiogenic cells and their arterial commitment via STAT5 activation. Blood, 2008,112(2):350–361
Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev, 2008,22(6):711–721
Hosui A, Kimura A, Yamaji D, et al. Loss of STAT5 causes liver fibrosis and cancer development through increased TGF-ta and STAT3 activation. J Exp Med, 2009,206(4):819–831
Yang X, Qiao D, Meyer K, et al. Signal transducers and activators of transcription mediate fibroblast growth factor-induced vascular endothelial morphogenesis. Cancer Res, 2009,69(4):1668–1677
Freie BW, Eisenman RN. Ratcheting Myc. Cancer Cell, 2008,14(6):425–426
Li Z, Boone D, Hann SR. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad Sci USA, 2008,105(48):18794–18799
Acosta JC, Ferrándiz N, Bretones G, et al. Myc inhibits p27-induced erythroid differentiation of leukemia cells by repressing erythroid master genes without reversing p27-mediated cell cycle arrest. Mol Cell Biol, 2008,28(24):7286–7295
Xu XL, Fang Y, Lee TC, et al. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell, 2009,137(6): 1018–1031
Ceballos E, Munoz-Alonso MJ, Berwanger B, et al. Inhibitory effect of c-Myc on p53-induced apoptosis in leukemia cells. Microarray analysis reveals defective induction of p53 target genes and upregulation of chaperone genes. Oncogene, 2005,24(28):4559–4571
Author information
Authors and Affiliations
Additional information
This project was supported by a grant from the State Key Basic Research Program (No. 2002CB513100) and a grant from the National Natural Science Foundation of China (No. C03020705).
Rights and permissions
About this article
Cite this article
Hong, Z., Han, Z., Xiao, M. et al. Microarray study of mechanism of trichostatin a inducing apoptosis of Molt-4 cells. J. Huazhong Univ. Sci. Technol. [Med. Sci.] 29, 445–450 (2009). https://doi.org/10.1007/s11596-009-0411-y
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-009-0411-y