Abstract
A total of 159 colonies of Chalara fraxinea were isolated between 2005 and 2006 from dying trees of European ash (Fraxinus excelsior L.) aged between 3 and 10 years. They derived from five regions of Poland differing by geographic location and climatic conditions. On the basis of 90 RAMS markers, pathogen intra- and inter-population variability, as well as its dependency on geographic distance and climatic conditions in the regions of strain origin, was analysed. The applied measures of intrapopulation genetic variability (genetic distance, Nei’s unbiased diversity, Shannon’s Information Index and percentage of polymorphic loci) allowed for differentiation of two strain groups: the first deriving from lowlands and the second from uplands and mountainous areas. Strains in lowlands were characterised by smaller number of markers, smaller number of polymorphic loci and smaller intrapopulation genetic variability. Positive and statistically significant correlation was shown between variability of isolates and elevation of regions above sea level. Pair-wise genetic distances between groups of isolates (Nei’s unbiased genetic distance) from particular regions were not significantly correlated with the corresponding geographic distances. On the basis of AMOVA, it was shown that 85% of variability was within-region differences and 2% between-region differences, whereas differences between lowlands and uplands were 13%. Principal Components Analysis (PCA) for the investigated regions confirmed the results from Nei’s genetic distance matrix.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
At the beginning of the 1990s, the first symptoms of increased dieback of ash (Fraxinus excelsior L.) were observed in north-eastern Poland. In the subsequent years, the disease symptoms were observed in all regions of the country, both in lowlands and in the mountains (Kowalski 2001; Kowalski and Łukomska 2005; Przybył 2002). Ash trees also started to display dieback symptoms in many countries of northern, western and, recently, also southern Europe (Bakys et al. 2009b; Engesser et al. 2009; Halmschlager and Kirisits 2008; Ioos et al. 2009; Kowalski and Holdenrieder 2008; Lygis et al. 2005; Ogris et al. 2010; Szabo 2008; Talgo et al. 2009). Tree stands of all age classes are suffering from the disease, both of natural and artificial afforestation, regardless of the occupied habitat, in forests and in urban areas (Bakys et al. 2009b; Halmschlager and Kirisits 2008; Kowalski and Holdenrieder 2008; Schumacher et al. 2007a; Talgo et al. 2009). Sick trees display a wide range of symptoms: leaf necrosis, wilting, premature leaf shedding, cankers on shoots, branches and stems, wood discoloration, dying of whole branches or their apices, top dying, and dying of whole trees and stands (Bakys et al. 2009a; Engesser et al. 2009; Kirisits and Halmschlager 2008; Kowalski 2006; Kowalski and Holdenrieder 2008; Schumacher et al. 2007b; Skovsgaard et al. 2010; Thomsen et al. 2007).
Numerous potentially pathogenic fungi from the genera Cytospora, Diplodia, Fusarium, Nectria and Phomopsis were identified in tissues of sick and decaying ash trees (Kowalski and Łukomska 2005; Przybył 2002; Schumacher et al. 2007b; Vasiliauskas et al. 2006). In Poland, a new species of anamorphic fungus, Chalara fraxinea T. Kowalski, was discovered on sick ash trees (Kowalski 2006). In the entire country, it is the most frequent fungus found on ash trees with dieback symptoms (Kowalski 2007, 2009). In recent years, its presence has been recorded in a majority of European countries where ash trees have started to die: in Lithuania, Austria, Germany, Sweden, Denmark, Finland, Norway, Czech Republic, Slovakia, Slovenia, Switzerland, France, Croatia, Hungary and Italy (Bakys et al. 2009a; Barić and Diminić 2009; Barklund 2006; Engesser et al. 2009; Halmschlager and Kirisits 2008; Ioos et al. 2009; Jankovsky and Holdenrieder 2009; Kowalski and Holdenrieder 2008; Ogris et al. 2010; Schumacher et al. 2007b; Szabo 2008; Thomsen et al. 2007). Pathogenicity of C. fraxinea with respect to Fraxinus excelsior has recently been demonstrated a number of times (Bakys et al. 2009b; Kirisits et al. 2009; Kowalski and Holdenrieder 2009a; Ogris et al. 2010; Talgo et al. 2009). Taking into account the significant economic and ecological threat, C. fraxinea was included in the EPPO Alert List and in the NAPPO Phytosanitary Alert System.
Recently, its teleomorph has been detected and assigned to Hymenoscyphus albidus (Roberge ex Desm.) W. Phillips (Kowalski and Holdenrieder 2009b), which had been known from Europe since 1851 as a saprotroph, not causing ash tree disease (Desmazières 1851; Ellis and Ellis 1997). It produces abundant apothecia on previous year ash leaf rachises in the litter, while ascospores are distributed in a period from July to September. Further research provided molecular evidence for the existence of, apart from H. albidus s. str., another morphologically similar fungus which was described as a new cryptic species Hymenoscyphus pseudoalbidus. According to the data collected so far, H. pseudoalbidus occurs on Fraxinus excelsior with disease symptoms, whereas H. albidus is present in ash tree stands without dieback (Queloz et al. 2010).
Data on genetic variability provide valuable insight in dispersal mechanisms and local adaptation of pathogens. We studied the genetic variability of Ch. fraxinea isolates from necrotic lesions on ash trees in different regions of Poland.
Materials and methods
Isolates
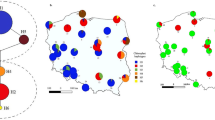
The research encompassed 159 isolates of Chalara fraxinea obtained between 2005 and 2006 from local necroses on live shoots or dead shoots of Fraxinus excelsior (Table 1). They derived from five regions of Poland, where disease symptoms occurred on young, 3- to 10-year-old ash tree stands. In every region, isolates derived from 2–4 Forest Districts (Table 1, Fig. 1). One region represented northern Poland (Region I), one eastern Poland (Region II), whereas three regions represented southern Poland (Region III, IV and V). Regions differed by geographic location and elevation above sea level, as well as climatic conditions, such as average annual temperatures, annual sum of precipitation and number of days with snow cover (Table 2).
Fungi isolation was performed during 24 (as an exception 48) h from collection of shoots in the field. From each shoot, a section whose length ranged between 8 and 10 cm with symptoms of local necrosis was cut out or, in the case of decayed shoot tops, from a section bordering on a live shoot. After superficial disinfection (1 min ethanol 96%, 5 min NaOCl 4%, 30 s ethanol 96%), parts of shoots whose dimensions were 5 × 2 × 2 mm were cut out and placed on the surface of 2% of malt extract agar (MEA; 20 g l-1 malt extract (Difco; Sparks, MD, USA), 15 g l-1 agar Difco supplemented with 100 mg l-1 streptomycin sulphate) in Petri dishes. From every shoot, 6–12 fragments were isolated. Incubation was conducted in the dark at room temperature. Among many colonies of various fungi, between 1 and 12 colonies of Ch. fraxinea appeared. Among them, 1 colony was selected at random. The growing mycelium was transferred to new MEA plates and incubated at 20°C in the dark.
Molecular analyses
Extraction of genomic DNA was conducted in line with the procedure of Carlson et al. (1991). Ribosomal DNA fragments containing ITS1, 5.8 S and ITS2 were amplified using ITS1 and ITS4 primers (White et al. 1990). PCR reaction was conducted in a volume of 50 μl containing: PCR buffer, MgCl2 1.5 mM, dNTP 200 mM (Fermentas, Burlington, Canada), primer 1 μM, Taq DNA polymerase 1 U (Qiagen, Valencia, CA, USA), template DNA 10 ng. PCR was conducted in Biometra T3 thermocycler (Biometra, Goettingen, Germany) programmed as follows: initial denaturation at 95°C for 5 min, subsequently 36 cycles consisting of denaturation at 95°C for 1 min, annealing for 45 s at 54°C, elongation at 72°C for 2.5 min. Elongation of the last cycle was extended to 8 min. Purification and sequencing of amplification products using ITS1 primer were carried out at Macrogen Europe, Netherlands. BLAST searches were conducted using Geneious 5.1 software (Biomatters, New Zeland) (Drummond et al. 2010).
Genetic polymorphism of isolates was determined by RAMS method (Random Amplified Microsatellites) (Hantula et al. 1996; Zietkiewicz et al. 1994). Eight primers were tested, among which most relevant ones were selected on the basis of electrophoretic profiles and repeatability of results (Table 3). DNA amplification of fungus was conducted in 10 μl of reactive mixture consisting of: PCR buffer, MgCl2 1 mM, dNTP 200 mM, 1 μM primer, Taq DNA polymerase 0.2 U, DNA 10 ng. Cycling parameters were: initial denaturation at 95°C for 5 min, subsequently 36 cycles consisting of denaturation at 95°C for 1 min, annealing for 45 s at temperatures depending on the type of primer (Table 3), elongation at 72°C for 2.5 min. Elongation of the last cycle was extended to 8 min. Amplification products were resolved in 1.5% agarose gel (Prona, Madrid. Spain) containing 0.2 μg/ml of ethidium bromide in the 1 × TBE buffer. GeneRuler 100 bp Ladder Plus (Fermentas) was used as a marker for the length of DNA fragments. The lengths of amplified products were determined using BIO 1 D++ software (Vilber Lurmat, Torcy, France). Every PCR reaction was performed twice for the purpose of confirming repeatability of results.
Data analysis
The RAMS bands that could be scored unequivocally were included in the analysis. They were scored as present (1) or absent (0) to create a binary matrix. Homogeneity in distribution frequency of allele for isolates deriving from individual regions was ascertained on the basis of the conducted chi-squared test. Binary data were used for the calculation of Dice’s similarity coefficient SD = [2a/(2a + b + c)], where a is the number of bands common for isolate x and y, b is the number of bands present only in isolate x, and c is the number of bands present only in isolate y. Similarity coefficient matrix was calculated with the use of SPSS Statistics 17.0 (SPSS, Chicago, Il, USA).
The intrapopulation genetic variation analysis between isolates obtained from particular regions was quantified on the basis of several measures: (1) genetic distance calculated as D = 1 – SD; (2) mean of Nei’s unbiased diversity (Hedrick 2000; Nei 1972); (3) Shannon’s Information Index (Lewontin 1972; Shannon and Weaver 1949); and (4) percentage of polymorphic loci. Calculations of these diversity estimates were performed with Popgene version 1.32 (Yeh et al 1997) and GenAlex 6.3 (Peakall and Smouse 2006) software.
Analysis of molecular variance (AMOVA) was carried out in GeneAlex 6.3 to study the occurrence of population structure. In this analysis, the total variation in the marker frequencies was divided into within- and between-region components. Variability components were expressed as a percentage of the entire variability, whereas statistical significance of components was tested with the use of 1,000 permutations. Genetic distance between groups of isolates from the analysed regions was expressed by means of Nei’s unbiased genetic distance (Hedrick 2000; Nei 1972).
The Principal Components Analysis (PCA), which graphically presents between-region variability, was performed based on genetic distance between region pairs, calculated according to Huff et al. (1993) method. The above-mentioned statistical analyses were performed with the GeneAlex 6.3 (Peakall and Smouse 2006) software.
In order to determine dependency of climatic conditions (Table 2) of studied regions on intrapopulation measures of genetic variability, correlation ratios were calculated using Statistica 8.0 (Stat Soft, Tulsa, OK, USA).
Results
On the basis of morphological researches and analysis of ribosomal DNA sequences containing ITS1, 5.8 S and ITS2 we identified 159 isolates of Chalara fraxinea. Analysed sequences were closest to Chalara fraxinea sequence No. FJ429385 (percent of identical sites: 99.6–100%).
On the basis of repeatability of results and the degree of polymorphism of electrophoretic profiles, among eight tested RAMS primers, the five most appropriate ones were selected (Table 3). The applied primers allowed for obtaining 90 amplification products in total. The example gel image, showing the banding pattern obtained using the AGC primer for regions I and V, is presented in Fig. 2. The number of obtained amplicons and the range of their length depended on the applied primer and the region of isolates origin (Tables 3 and 4). None of the obtained amplification product (RAMS marker) was monomorphic for the group of isolates deriving from individual regions. Similarly, no pair of isolates with the same haplotype was ascertained for the applied primers. Chi-squared test showed that the obtained data were homogenous (the homogeneity test was insignificant at P < 0.01), which allowed for the application of variance analysis (ANOVA) for evaluation of differences between the mean genetic distance (D) values for the regions.
On the basis of applied measures of intrapopulation genetic variability, the analysed regions of occurrence of Chalara fraxinea were divided into two groups. The first one encompassed lowland regions (Region I and Region II), whereas the second one highland regions (Region III and Region IV) and a mountainous region (Region V). Isolates derived from lowland regions were characterised by a lower number of obtained amplification products and a smaller number of polymorphic loci, as well as smaller values of measures of intrapopulation genetic variability. All the above-listed indices were lower by over 60% in the case of lowland regions in comparison to regions located higher up (Table 4). Additional cluster analyses for each individual region and for the combined low and high elevation regions revealed the presence of several genetically distinct groups, which, however, were not related to the geographic origin of the specimens (data not shown).
The applied measures of intrapopulation genetic variability of isolates within regions were significantly correlated (Table 5). Genetic distance among isolates in individual regions increased along with growth of elevation of a given region above sea level (Tables 2 and 4). A positive, statistically significant correlation between measures of intrapopulation genetic variability of isolates and elevation of regions above sea level was ascertained (Table 5). A scatter plot of pair-wise genetic distance (D) and average elevation of regions illustrated a gradual increase in genetic distance between regions at increasing elevations (Fig. 3). The R 2 value (0.897) suggested that almost 90% of total variation in genetic distance was explained by changes in elevations between regions. It was also ascertained that genetic distance (D) was significantly correlated with the number of days with snow cover in the examined regions (Table 5).
Out of 90 RAMS markers obtained, 7 occurred with a frequency decreasing along with increase in the region’s elevation above sea level. They were obtained with the use of primers: ATG (ATG_1259 bp), AGT (AGT_650), AGC (AGC_1738, AGC_1216, AGC_1080, AGC_490) and ACA (ACA_643). Five of them showed statistically significant, negative correlation with the elevation of a region above sea level (Table 6).
Genetic distance among strain groups deriving from individual regions was calculated with the use of unbiased Nei’s genetic distance (Table 7). The matrix of pair-wise genetic distances between regions was not significantly correlated with the corresponding matrix of geographic distances (Mantel tests; r = 0.418, P = 0.133; Fig. 4). Regions located lowest (I and II) were characterised by smallest genetic distance, in spite of being most distanced geographically. Pairs of regions that were geographically close were most distanced genetically—they were located higher above sea level (regions III, IV and V) and characterised by greater intrapopulation variability (Tables 2 and 4). Correlation analysis has shown that Nei’s genetic distance was significantly correlated with differences in elevation above sea level (Pearson rank correlation coefficient: r = 0.769, P = 0.009) and differences in measures of intrapopulation variability of isolates for regions of their occurrence (Tables 4, 7).
The AMOVA of distance matrix for the five investigated regions permitted partitioning of the overall variation into three levels. The proportion of variation attributable to within-region differences was 85% (P = 001), between-region differences only 2% (P = 0.005), whereas differences between lowlands (Regions I and II) and uplands (Regions III–V) were 13% (P = 0.001) (Table 8).
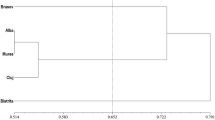
Principal Components Analysis (PCA) for investigated regions confirmed the results from Nei’s genetic distance matrix (Table 7). A close relationship was revealed between groups of isolates from lower placed Regions I and II, and between three regions from southern Poland, which are located higher. It should be stressed that the most distanced group of isolates was obtained from Region V, which is placed on the highest elevation (Fig. 5).
Discussion
The study describes the genetic variability of Ch. fraxinea, a serious pathogen of ash trees. The applied molecular markers showed a high genetic variability and an interregional genetic structure of the fungal populations, depending on geographic location and climatic conditions. RAMS markers are frequently used to characterise the genetic variability because they are highly polymorphic and, for the analysis, only a small amount of mycelium is needed (Grünig et al. 2001; Kraj and Kowalski 2008; McDonald 2004; Vainio and Hantula 1999).
The genetic variability of Ch. fraxinea was determined at multiple loci across the genome. A potentially more sensitive measure of genetic variability is determination of the number of allele on a locus; however, this measure cannot be applied in this case.
We did not find individuals with identical haplotypes. This leads to the conclusion that the only method of Ch. fraxinea propagation is creation of ascospores. It was observed by Kirisits et al. (2009) who showed that conidia did not germinate on MEA, V8 agar or on agar medium containing an extract from ash leaves that stimulated mycelial growth. Also, Rytkönen et al. (2010) observed a large proportion of haplotypes (14 among 32 isolates) and high genetic variability of fungus.
Among the most important sources of genetic variability of fungi are: selection of various alleles and genotypes, depending on the local environment, limitations of gene flow and founder effects and epidemic spread to new host plant populations (Hedrick 2000; Wang et al. 1997). All the measures of intrapopulation variability applied in our study showed that genetic variability is positively correlated with elevation above sea level. Comparison of intrapopulation genetic variability among isolates from lowlands and areas at higher elevations revealed clear differences. No specific molecular markers characteristic for distinguished groups of regions (lowlands and uplands) were identified, yet for five markers a statistically significant, negative correlation between their frequency and increasing elevation of the region’s location above sea level was shown (Table 6). According to our data, the genetic variability of Ch. fraxinea isolates is not connected to the geographic distance of regions of their occurrence, as it has been found for other fungi. For example, the main factors influencing genetic variability of Gremmeniella abietina (Lagerb.) Morelet are climatic conditions and thickness of snow cover (Kraj and Kowalski 2008; Uotila et al. 2006). Higher levels of genetic variability of isolates from sites at higher elevations were related to the more harsh and variable climate. Strains of Sclerophoma pythiophila (Corda) v. Hoehn deriving from various regions also displayed high level of intrapopulation variability, dependant on climatic conditions (Kraj 2009). For Ch. fraxinea in Poland, the effect of climatic conditions on the level of intrapopulation variability was confirmed by AMOVA analysis (Table 8), which showed a much higher (13%) share of variability among isolates deriving from lowlands and uplands in comparison to only 2% share in variability between geographically distant regions. This means that 87% of the differences can be explained by elevation. We assume that the high genetic variability level of Ch. fraxinea is related to the need for adaptation to climate. Development and spreading of fungal pathogens depend strongly on humidity and temperature and coping with these factors is essential for survival (Huber and Gillespie 1992). The high genetic variability of Ch. fraxinea populations, which possibly evolved in response to climatic conditions, could also contribute to virulence. Isolates of Ch. fraxinea, depending on temperature, showed a significant variety of the growth rate in vitro (Kowalski and Bartnik 2010). Such growth differences occurred not only among isolates from distant origins, but also among isolates deriving from the same forest district. Inoculation experiments showed tolerance of individual ash trees against Ch. fraxinea both in the greenhouse (Bakys et al. 2009a) and in the field (Olrik et al. 2007).
High levels of genetic variation are characteristic for sexually reproducing organisms with a wide geographical distribution (Hedrick 2000; James et al. 1999) and the contrary is true for species with a restricted distribution, small populations or in organisms that reproduce asexually (Burdon and Roelfs 1985). The high genetic variability is somewhat surprising for a pathogen, which has probably been introduced in Europe (Queloz et al. 2010). All strains in our study were clearly determined as Ch. fraxinea with the teleomorph H. pseudoalbidus by ITS sequencing. Nevertheless, we cannot exclude completely that cryptic species might be detected within this taxon by application of markers with higher resolution (e.g. AFLPs).
References
Bakys R, Vasaitis R, Barklund P, Ihrmark K, Stenlid J (2009a) Investigations concerning the role of Chalara fraxinea in declining Fraxinus excelsior. Plant Pathol 58:284–292
Bakys R, Vasaitis R, Barklund P, Thomsen IM, Stenlid J (2009b) Occurrence and pathogenicity of fungi in necrotic and non-symptomatic shoots of declining common ash (Fraxinus excelsior) in Sweden. Eur J For Res 128:51–60
Barić L, Diminić D (2009) First report of the pathogenic fungus Chalara fraxinea Kowalski on common ash (Fraxinus excelsior L.) in Gorski Kotar. Glas biljn zašt 10:33
Barklund P (2006) Okänd svamp bakom askskottsjukan. Värsta farsoten som drabbat en enskild trädart (Unknow fungus causes dieback of ash shoots). SkogsEko 3:10–11
Burdon JJ, Roelfs AP (1985) The effect of sexual and asexual reproduction on the isozyme structure of population Puccinia graminis. Phytopathology 75:1068–1073
Carlson JE, Tulsieram LK, Glaubitz JC, Luk VWK, Kauffeldt C, Rutledge R (1991) Segregation of random amplified DNA markers in F1 progeny of conifers. Theor Appl Genet 83:194–200
Desmazières JB (1851) Peziza (Phialea cyathoidea) albida. Ann Sci Nat, Bot Ser 3, Tome 16, 823–824
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v5.1
Ellis MB, Ellis JP (1997) Microfungi on land plants. An identification handbook. Richmond Slough
Engesser R, Queloz V, Meier F, Kowalski T, Holdenrieder O (2009) Das Triebsterben der Esche in der Schweiz. Wald und Holz 6:24–27
Grünig CR, Sieber TN, Holdenrieder O (2001) Characterization of dark septate endophytic fungi (DSE) using inter-simple-sequence repeat-anchored polymerase chain reaction (ISSR-PCR) amplification. Mycol Res 105:24–32
Halmschlager E, Kirisits T (2008) First report of Chalara fraxinea on Fraxinus excelsior in Austria. New Dis Rep 17:20. [http://www.ndrs.org.uk/article.php?file=2008-25.asp]
Hantula J, Dusabenyagasani M, Hamelin RC (1996) Random amplified microsatellites (RAMS) – a novel method for characterizing genetic variation within fungi. Eur J For Pathol 26:159–166
Hedrick PW (2000) Genetics of populations, 2nd edn. Jones and Bartlet, Boston, MA
Huber L, Gillespie TJ (1992) Modeling leaf wetness in relation to plant disease epidemiology. Annu Rev Phytopathol 30:553–577
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss Buchloe dactyloides (Nutt) Engelm. Theor Appl Genet 86:927–934
Ioos R, Kowalski T, Husson C, Holdenrieder O (2009) Rapid in planta detection of Chalara fraxinea by a real − time PCR assay using a dual-labelled probe. Eur J Plant Pathol 125:329–335
James TY, Porter D, Hamrick JL, Vilgalys R (1999) Evidence for limited intercontinental gene flow in the cosmopolitan mushroom, Schizophyllum commune. Evolution 53:1665–1667
Jankovsky L, Holdenrieder O (2009) Chalara fraxinea – ash dieback in the Czech Republic. Plant Prot Sci 45:74–78
Kirisits T, Halmschlager E (2008) Eschenpilz nachgewiesen. Forstztg 119:32–33
Kirisits T, Matlakova M, Mottinger-Kroupa S, Cech T, Halmschlager E (2009) The current situation of ash dieback caused by Chalara fraxinea in Austria. In: Doğmuş-Lehtijärvi T (ed) Proceedings of the conference of IUFRO working party 7.02.02, Eğirdir, Turkey, 11-16 May 2009. SDU Faculty of Forestry Journal, Serial: A, Special Issue: pp 97-119
Kowalski T (2001) O zamieraniu jesionów [About ash dieback]. Trybuna Leśnika 4:6–7
Kowalski T (2006) Chalara fraxinea sp. nov. associated with dieback of ash (Fraxinus excelsior) in Poland. For Pathol 36:264–270
Kowalski T (2007) Chalara fraxinea – new described fungus species on dying ash in Poland. Sylwan 4:44–48
Kowalski T (2009) Rozprzestrzenienie grzyba Chalara fraxinea w aspekcie procesu chorobowego jesionu w Polsce (Expanse of Chalara fraxinea fungus in terms of ash dieback in Poland). Sylwan 10:668–674
Kowalski T, Bartnik C (2010) Morphological variation in colonies of Chalara fraxinea isolated from ash stems with symptoms of dieback and effects of temperature on colony growth and structure. Acta Agrobot 63:99–106
Kowalski T, Holdenrieder O (2008) Eine neue Pilzkrankheit an Esche in Europa. Schweiz Z Forstwes 159:45–50
Kowalski T, Holdenrieder O (2009a) Pathogenicity of Chalara fraxinea. For Pathol 39:1–7
Kowalski T, Holdenrieder O (2009b) The teleomorph of Chalara fraxinea, the causal agent of ash dieback. For Pathol 39:304–308
Kowalski T, Łukomska A (2005) Badania nad zamieraniem jesionu (Fraxinus excelsior L.) w drzewostanach Nadleśnictwa Włoszczowa (Study on ash dying in the Włoszczowa Forest Unit stands). Acta Agrobot 59:429–440
Kraj W (2009) Differentiation and genetic structure of Sclerophoma pythiophila strains on Pinus sylvestris in Poland. J Phytopathol 157:400–406
Kraj W, Kowalski T (2008) Genetic variation in Polish strains of Gremmeniella abietina. For Pathol 38:203–217
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:381–394
Lygis V, Vasiliauskas R, Larsson K, Stenlid J (2005) Wood-inhabiting fungi in stems of Fraxinus excelsior in declining ash stands of northern Lithuania, with particular reference to Armillaria cepistipes. Scand J For Res 20:337–346
McDonald BA (2004) Population Genetics of Plant Pathogens. Plant Health Instructor. doi:10.1094/PHI-A-2004-0524-01
Nei M (1972) Genetic distance between populations. Am Nat 106:283–392
Ogris N, Hauptman T, Jurc D, Floreancig V, Marsich F, Montecchio L (2010) First report of Chalara fraxinea on common ash in Italy. Plant Dis 94:133
Olrik DC, Kjaer ED, Ditlevsen B (2007) Clonal differences in attacks by ash shoot dieback. Skoven 39:522–525
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Przybył K (2002) Fungi associated with necrotic apical parts of Fraxinus excelsior shoots. For Pathol 32:387–394
Queloz V, Grünig CR, Berndt R, Kowalski T, Sieber TN, Holdenrieder O (2010) Cryptic speciation in Hymenoscyphus albidus. For Pathol 40 (in press). doi:10.1111/j.1439-0329.2010.00645.x
Rytkönen A, Lilja A, Drenkhan R, Gaitnieks T, Hantula J (2010) First record of Chalara fraxinea in Finland and genetic variation among isolates sampled from Ǻland, mainland Finland Estonia and Latvia. For Pathol. doi:10.1111/j.1439-0329.2010.00647.x
Schumacher J, Heydeck P, Leonhard S, Wulf A (2007a) Neuartige Schaeden an Eschen. Allg Forst Z – Der Wald 62, 20:1094–1096
Schumacher J, Wulf A, Leonhard S (2007b) Erster Nachweis von Chalara fraxinea T. Kowalski sp. nov., in Deutschland – ein Verursacher neuartiger Schaeden an Eschen. Nachr Deutsch Pflanzenschutzd 59:121–123
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Skovsgaard JP, Thomsen IM, Skovgaard IM, Martinussen T (2010) Associations between symptoms of dieback in even-aged stands of ash (Fraxinus excelsior L.). For Pathol 40:7–18
Szabo I (2008) First report of Chalara fraxinea affecting common ash in Hungary. New Dis Rep 18:30 [http://www.ndrs.org.uk/article.php?id=018030]
Talgo V, Sletten A, Brurberg MB, Solheim H, Stensvand A (2009) Chalara fraxinea isolated from diseased ash in Norway. Plant Dis 93:548
Thomsen IM, Skovsgaard JP, Barklund P, Vasaitis R (2007) Svampesygdom er arsag til toptorre i ask. [Fungal disease is the cause of ash dieback]. Skoven 39:234–236
Uotila A, Kurkela T, Tuomivirta T, Hantula J, Kaitera J (2006) Gremmeniella abietina types cannot be distinguished using ascospore morphology. For Pathol 36:395–405
Wang X, Ennos RA, Szmidt AE, Hansson P (1997) Genetic variability in the canker pathogen fungus, Gremmeniella abietina. 2. Fine-scale investigation of the population genetic structure. Can J Bot 75:1460–1469
White TJ, Bruns TD, Lee S, Taylor J, (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: A Guide to Methods and Applications. Academic, New York, pp 315–322
Vainio EJ, Hantula J (1999) Variation of RAMS markers within the intersterility groups of Heterobasidion annosum in Europe. Eur J For Pathol 29:231–246
Vasiliauskas R, Bakys R, Lygis V, Ihrmark K, Barklund P, Stenlid J (2006) Fungi associated with the decline of Fraxinus excelsior in the Baltic States and Sweden. In: Oszako T, Woodward S (eds) Possible limitation of decline phenomena in broadleaved stands. Forest Research Institute, Warsaw, pp 45–53
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1997) POPGENE. Molecular Biology and Biotechnology Center, University of Alberta, Canada, The User Friendly Software for Population Genetic Analysis
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR) anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgement
This study was financially supported by The Ministry of Science and Higher Education, grant No. N N309 146937
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Kraj, W., Zarek, M. & Kowalski, T. Genetic variability of Chalara fraxinea, dieback cause of European ash (Fraxinus excelsior L.). Mycol Progress 11, 37–45 (2012). https://doi.org/10.1007/s11557-010-0724-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-010-0724-z