Abstract
Objective
To calculate by means of a computer-aided tool the volumes of healthy residual lung parenchyma, of emphysema, of ground glass opacity (GGO) and of consolidation on chest computed tomography (CT) in patients with suspected viral pneumonia by COVID-19.
Materials and methods
This study included 116 patients that for suspected COVID-19 infection were subjected to the reverse transcription real-time fluorescence polymerase chain reaction (RT-PCR) test. A computer-aided tool was used to calculate on chest CT images healthy residual lung parenchyma, emphysema, GGO and consolidation volumes for both right and left lung. Expert radiologists, in consensus, assessed the CT images using a structured report and attributed a radiological severity score at the disease pulmonary involvement using a scale of five levels. Nonparametric test was performed to assess differences statistically significant among groups.
Results
GGO was the most represented feature in suspected CT by COVID-19 infection; it is present in 102/109 (93.6%) patients with a volume percentage value of 19.50% and a median value of 0.64 L, while the emphysema and consolidation volumes were low (0.01 L and 0.03 L, respectively). Among quantified volume, only GGO volume had a difference statistically significant between the group of patients with suspected versus non-suspected CT for COVID-19 (p < < 0.01). There were differences statistically significant among the groups based on radiological severity score in terms of healthy residual parenchyma volume, of GGO volume and of consolidations volume (p < < 0.001).
Conclusion
We demonstrated that, using a computer-aided tool, the COVID-19 pneumonia was mirrored with a percentage median value of GGO of 19.50% and that only GGO volume had a difference significant between the patients with suspected or non-suspected CT for COVID-19 infection.
Similar content being viewed by others
Introduction
The novel coronavirus that was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses caused a pneumonia outbreak worldwide [1,2,3,4]. The disease caused by the virus, named coronavirus disease (COVID-19), can be spread through human-to-human contact.
In Italy, the geographical distribution of the infection was uneven, with the northern regions that face disproportionately higher numbers of contagions and deaths compared with the central and southern regions [5]. To support Italian radiologists in becoming familiar with COVID-19 imaging features, the Italian Society of Medical and Interventional Radiology (SIRM) established a dedicated page on the SIRM Web site with an Italian collection of COVID-19 examinations and provided operational procedures to help radiology units in the daily management of COVID-19 patients [5].
The estimation of the incubation period mean value was of 5.2 days, which allows air travelers to spread the disease globally [6, 7]. Evidence shows that virus transmission can occur during the incubation period in asymptomatic patients. The COVID-19 diagnosis is confirmed by the positive results of the nucleic acid amplification test of the respiratory tract or blood specimens using reverse transcription real-time fluorescence polymerase chain reaction (RT-PCR).
In the context of the COVID-19 pandemic, there is a scrupulous need for ready-to-use resources for data acquisition, the computerized tool and artificial intelligence (AI) algorithms to speed up the search for effective and safe treatments. The progressive integration of AI-based solutions in healthcare is already changing established paradigms in the entire healthcare ecosystem, leveraging the progressive digitalization of medical data. In particular, diagnostic and decision support systems developed for medical imaging are the first successful examples of innovation for health. Radiomics and AI-based methods have led to diagnostic applications that accelerate image acquisition, preprocessing, annotation and interpretation, offering an “augmentation” of the radiologists, rather than their unrealistic substitution. In particular, the application of AI in medical imaging has improved the assessment, diagnosis and early detection of neurodegenerative diseases and heart diseases, with specifically high impact on breast and lung cancer.
Medical imaging has been efficiently applied for diagnostics and management of COVID-19 pneumonia. Mainly, computed tomography (CT) examination has been used extensively in China and now worldwide to evaluate the grade and the extension of the viral pneumonia by COVID-19 and in the follow-up, also based on AI algorithms [8,9,10,11,12]. The most Italian hospitals are employing chest X-ray as the first-line method, with faster results comparing with those of RT-PCR, especially by using portable X-ray units which reduce the movement of patients and so minimizing the risk of cross-infection [10].
Several radiological organizations do not recommend CT as a primary diagnostic tool for COVID-19 viral infection [13,14,15,16,17]. However, the diagnosis of viral pneumonia based on chest CT may be used to recommend patient isolation. Further, chest CT has an important role in the management of patients with suspected SARSCoV-2 infection, identifying patients with severe lung involvement of viral pneumonia at CT; this could direct the clinician versus an anticipation of mild invasive ventilation that has been demonstrated effective in reducing severity of pneumonia [18,19,20,21,22,23,24,25,26,27]. The main CT characteristics reported in the recent literature and individuated in patients affected by COVID-19 pneumonia were ground-glass opacity (GGO), consolidation, reticulation/thickened interlobular septa, nodules and lesion distribution in left, right or bilateral lungs [17,18,19].
We analyzed the chest CT images performed at the time of admission of 116 patients with suspected SARS-CoV-2 infection in order to calculate automatically by post-processing automatic tool the volume of healthy lung parenchyma, emphysema volume, GGO and consolidation volume. End-point of the study is also to identify the CT patterns linked and correlate with the severity of the COVID-19 viral infection.
Materials and methods
Patient characteristics
Institutional review board of “Bergamo Est” waived written informed consent for this retrospective study considering the COVID-19 emergency. The cohort was composed of 116 patients (32 women and 77 men; 68.8 years of mean age; range, 29–93 years) subjected to the nucleic acid amplification test of the respiratory tract or blood specimens using reverse transcription real-time fluorescence polymerase chain reaction test, between February 23, 2020, and March 5, 2020. The RT-PCR test was executed in the clinical laboratory of ASST Bergamo Est (Seriate, Italy).
CT technique
Chest CT scan was acquired at the time of patient admission in hospital with a 128 slice CT scanner (Ingenuity of Philips, Amsterdam, The Netherlands). CT examinations were performed with the patient in the supine position in breath hold during an inspiration using a standard dose protocol, without contrast intravenous injection. The scanning range was from the apex to the base of the lungs. The tube voltage and the current tube were 120 kV and 100–200 mA, respectively. All data were reconstructed with 0.6–1.0 mm increment. The matrix was 512 × 512 mm2. Images were reconstructed using a sharp reconstruction kernel for parenchyma and viewed at window settings optimized for the assessment of the lung parenchyma (window width, 1600 HU; window level, 600 H.U).
Post-processing with thoracic VCAR software
DICOM data were transferred into a PACS workstation (AW SERVER 3.2 ext 3.0, of GE Healthcare, Chicago, Illinois, USA), and CT images were evaluated using a clinically available artificial intelligence computer tool, the Thoracic VCAR software (GE Healthcare, Chicago, Illinois, USA) in order to evaluate thoracic disease.
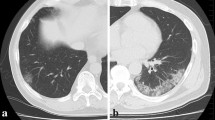
The software has the CE certification to document emphysema. The software provides automatic segmentation of the lungs and automatic segmentation and tracking of the airway tree. It provides the classification of voxels based on Hounsfield Units and a color-coded display of the segmented regions. We analyzed the CT scans by pre-setting a threshold value of Hounsfield Units in order to obtain a quantification for both lungs of emphysema ( − 1024/ − 977; blue) [20], of healthy residual lung parenchyma ( − 977/ − 703; yellow) [21, 22], of GGO ( − 703/ − 368; pink) and of consolidation (− 100/5; red) [21, 23,24,25] (Fig. 1). Chest CT evaluation using this computer tool was done on 116/134 (86.6%) patients because the software in 18 cases was not able to provide automatically quantification of thoracic disease.
Example of lung volumes calculated by computer-aided tool on CT images by pre-setting a threshold value of Hounsfield Units and a color in order to obtain a segmentation of both lungs and a quantitative evaluation of emphysema (− 1024/ − 977; blue), Healthy lung parenchyma (− 977/ − s703; yellow), GGO (− 703/ − 368; pink), consolidation (− 100/5; red) and other (− 368/ − 100; green)
Radiologist analysis
Radiologists attributed to each lung looking the CT images at the pulmonary involvement by disease a severity score using a scale of five levels (none, mild: ≤ 25% of involvement, moderate: 26–50% of involvement, severe: 51–75% of involvement and critic involvement: 76–100%). An overall radiological severity score was obtained summing the scores for each lung and then considering a mild severity a score ≤ 2, moderate 3–4, severe 5–6 and critic 7–8. Two radiologists with more than 10 years of thoracic-imaging analysis experience evaluated the severity of images in a double-blind manner. Another, more experienced, radiologist resolved any disagreement between the two radiologists.
Moreover, all chest CT examinations were reviewed using a structured report realized by Italian Society of Medical Radiology and Interventional Radiology (SIRM, Milan, Italy) in collaboration with the Exprivia Healthcare Company (Bari, Italy) [26]. The presence of GGO and consolidation was assessed in order to define their localization (unilateral and bilateral), axial distribution (diffuse, predominantly peripheral, mainly central, declivous, anti-declivous), distribution on the cranio-caudal plane (diffuse, multifocal/patching, prevalent in the upper lobes, prevalent in the lower lobes, gravitational) and their site.
Statistical analysis
Continuous data were expressed in terms of median value and range. Mann–Whitney test was performed to verify differences statistically significant between groups of continuous variables. Kruskal–Wallis test and chi-square test with Yates correction were used to assess statistically significant differences between percentage values.
p value < 0.05 was considered statistically significant for all tests.
All analyses were performed using IBM SPSS Statistics 24 (IBM, USA).
Results
CT and RT-PCR performance results
The patients’ characteristics are reported in Table 1.
Median value of temporal difference between RT-PCR tests execution and CT scan was 0.18 days ± 2.0 days. CT was suspected for COVID-19 viral pneumonia in 109 of 116 (94.0%) patients, while COVID-19 was diagnosed at RT-PCR in 90 of the 116 (77.6%) patients.
CT volumes analysis
No differences of the value of volumes between right and left lung were identified (p value > 0.05 at Mann–Whitney test).
GGO was the most represented characteristic in suspected CT by COVID-19 (Table 2), it was present in 102/109 (93.6%) patients. GGO percentage volume was 19.50% with a median value of 0.64 L, statistically superior (p value < < 0.01 at Mann–Whitney test) with respect to the other volumes (emphysema volume (0.01 L) and consolidation volume (0.03 L)).
GGOs were predominantly bilateral in 89/102 (87.3%) patients with involvement in multiple lobes in 77/102 (75.5%) patients, distribution peripheral in 82/102 (80.4%) patients and multifocal/patching in 69/102 (67.7%) patients. Consolidations were found in 78/109 (71.6%) patients, predominantly bilateral in 68/78 (87.2%) patients, with involvement in multiple lobes in 53/78 (67.9%) patients, distribution peripheral in 67/78 (85.9%) patients and segmental in 33/78 (42.3%) patients.
Among quantified volumes, only GGO had a difference statistically significant between patients with suspected CT and non-suspected CT for COVID-19 (p value < < 0.01 at Mann–Whitney test, Table 2).
In Table 3, the percentage values of volumes of healthy residual lung parenchyma, emphysema, GGO and consolidations are reported considering the patients grouped for pneumonia radiological severity score.
Volume median values of healthy residual lung parenchyma were, respectively, of 93.8%, 91.1%, 77.8%, 66.0% and 60.0% for none, mild, moderate, severe and critical radiological score.
Volume median values of GGO were, respectively, of 4.1%, 6.6%, 16.0%, 26.8% and 31.2% for none, mild, moderate, severe and critical radiological score.
Volume median values of consolidations were, respectively, of 0.6%, 0.7%, 0.9%, 1.3% and 1.60% for none, mild, moderate, severe and critical radiological score.
There were differences statistically significant among the groups based on radiological severity score in terms of healthy residual parenchyma volume, of GGO volume and of consolidations volume with a p value < < 0.001 at Kruskal–Wallis test (Fig. 2).
Discussion and conclusions
Several recent publications have described CT imaging features of COVID-19, the evolution of these features over time and the performance of radiologists in order to distinguish COVID-19 from other viral infections [8, 11, 27,28,29,30,31]. These studies reported that COVID-19 often produces a CT pattern resembling organizing pneumonia, notably peripheral ground-glass opacities and nodular or mass-like GGO that are often bilateral and involvement in multiple lobe.
Numerous artificial intelligence software applications have recently been proposed for the recognition of lung lesions from COVID-19 on chest CT examination, many of which are Chinese [32,33,34]. However, these tools are not recognized as medical devices because CE marking is absent. Furthermore, they have been tested on thousands of cases of COVID-19 but not equally on as many cases of non-COVID-19 coronavirus, affecting their ability to make a differential diagnosis. In this context, the role of artificial intelligence remains limited to the numerical quantization of the lesions and their distribution. Therefore, the aid of these artificial intelligence software is limited in experimental protocols to evaluate the therapies effectiveness or to compare several CT examinations in search of findings that can anticipate the outcome of the disease and the prognosis.
Gozes et al. [34] used 2D and 3D deep learning models to explore AI tools in the CT images analysis for detection, quantification and tracking of COVID-19. A total of 106 COVID-19 chest CT scans (and 99 normal ones) were used to find potential COVID-19 CT features and to evaluate progression of the disease in patients over time. Proof of principle of diagnostic capability of deep learning methods from CT images to screen COVID-19 have been recently demonstrated by Wang et al. [34] on 1,119 CT images of pathogen-confirmed COVID-19 cases versus typical viral pneumonia. The accuracy and applicability of deep learning for screening COVID-19 by CT examinations have, however, been questioned, based on concerns of the radiologists’ association and given the impact of selection bias reported in first published results. At the same time, radiomics approaches can be usefully implemented, focusing on segmentation techniques of the lung parenchyma based on region growing techniques and on other radiomics COVID-19-specific feature and their use with machine learning such as support vector machines or random forests [35,36,37,38,39,40].
In clinical practice, lung recruitment is usually considered a useful strategy [41, 42]. For this reason, it has been suggested that the condition of patients with a high percentage of potentially recruitable lung is better than that of patients with a lower percentage of potentially recruitable lung, given the presence of similar degrees of lung injury.
Several methods of disease extent quantification at chest CT have been proposed, including the extent of emphysema, pulmonary fibrosis and acute respiratory distress syndrome (ARDS) [43,44,45,46].
Grassi et al. compared different commercial software applications in the quantification of pneumonia lesions in COVID-19 infection in order to stratify the patients based on the disease severity using chest CT images. They concluded that computer-aided pneumonia quantification could be an easy and feasible way to stratify COVID-19 cases according to severity [47].
CT score of the burden of lung disease was previously reported as a risk factor for mortality in ARDS [43]. A variable combination of CT signs and their distribution affects the degree of diagnostic confidence in the diagnosis of COVID-19 infection (respectively high, intermediate and low or alternative diagnosis) but does not correlate with a statistically significant difference in the outcome prediction of patients. Therefore, the objective quantification by a post-processing automatic tool of the volumes of healthy lung parenchyma, emphysema, GGO and consolidation could be useful in the COVID-19 diagnosis and could be used to differentiate the patients with severe lung involvement.
According to recent publications, in this study we quantified using a computer tool the abnormalities visible at the chest CT images (emphysema, GGO and consolidations) and we reported that GGO was the most represented characteristic in suspected CT by COVID-19 with a percentage volume value of 19.50% and a median value of 0.64 L. Among quantified volumes, only GGO volume had a difference statistically significant between the group of patients with suspected CT and non-suspected CT for COVID-19. GGO abnormality, also in our cohort, was predominantly bilateral (87.3%), with involvement in multiple lobe (75.5%) involvement, peripheral (80.4%) and multifocal/patching (67.7%) distribution.
Moreover, we reported that no differences between right and left volume lung could be observed, while there were differences statistically significant among the groups based on radiological severity score in terms of healthy residual lung parenchyma volume, of GGO volume and of consolidations volumes.
The main limitations of the present study are as follows: the nature retrospective and monocentric of the study conducted on a cohort of symptomatic hospitalized patients from an area of high epidemiological risk and with a high pretest probability of COVID-19 infection; too small comparison group represented by the patients with CT no suspected for COVID-19 viral pneumonia; and the absence of clinical data to correlate with radiological findings. However, a future endpoint of the study is the correlation of the radiological finding and clinical one using artificial intelligence tools.
In conclusion, we demonstrated that the COVID-19 was mirrored mainly by GGO abnormality and that this had a percentage value of 19.50% with respect to the total lung volume. Moreover, we observed that only the GGO volume had a difference significant between the patients with suspected or non-suspected for COVID-19 infection. We think that analysis of CT findings using a computer tool based on different thresholding Hounsfield Units setting could be able to identify pulmonary abnormalities and lung recruitment and we believe that knowledge of the percentage of potentially recruitable lung may be important for establishing the management of COVID-19 pneumonia.
Abbreviations
- AI:
-
Artificial intelligence
- COVID-19:
-
Coronavirus disease 19CT = Computed tomography
- GGO:
-
Ground glass opacity
- MERS:
-
Middle East respiratory syndrome
- RT-PCR:
-
Reverse transcription real-time fluorescence polymerase chain reaction
- SARS:
-
Severe acute respiratory syndrome
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- WHO:
-
World Health Organization
References
World Health Organization website. Middle East respiratory syndrome coronavirus (MERS-CoV). www.who.int/emergencies/mers-cov/en/. Accessed March 21, 2020.
World Health Organization website. Naming the coronavirus disease (COVID-2019) and the virus that causes it. www.who.int/emergencies/diseases/. Accessed March 21, 2020.
Giovanetti M, Angeletti S, Benvenuto D, Ciccozzi M (2020) A doubt of multiple introduction of SARS-CoV-2 in Italy: a preliminary overview. J Med Virol. https://doi.org/10.1002/jmv.25773
Wuhan Coronavirus (2019-nCoV) Global Cases (by Johns Hopkins CSSE). Case Dashboard. https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6. Accessed March 21, 2020.
Albano D, Bruno A, Bruno F, Calandri M, Caruso D, Clemente A, Coppolino P, Cozzi D, De Robertis R, Gentili F, Grazzini I, Jannonae ML, Liguori C, Natella R, Pace G, Posa A, Scalise P, Accarino B, Bibbolino C, Barile A, Grassi R, Messina C, Young SIRM Working Group (2020) Impact of coronavirus disease 2019 (COVID-19) emergency on Italian radiologists: a national survey. Eur Radiol 14:1–10
Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382(13):1199–1207
Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wölfel R, Hoelscher M (2020) Transmission of 2019-nCoV infection from an asymptomatic contact in germany. N Engl J Med 382(10):970–971
Li Y, Xia L (2020) Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol 4:1–7
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet 395(10223):497–506
Cozzi D, Albanesi M, Cavigli E, Moroni C, Bindi A, Luvarà S, Lucarini S, Busoni S, Mazzoni LN, Miele V (2020) Chest X-ray in new coronavirus disease 2019 (COVID-19) infection: findings and correlation with clinical outcome. Radiol Med 125(8):730–737
Lei J, Li J, Li X, Qi X (2020) CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology 295(1):18
Novel Coronavirus Pneumonia Emergency Response Epidemiology Team (2020) The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 41(2):145–151
American College of Radiology. ACR Recommendations for the Use of Chest Radiography and Computed Tomography (CT) for Suspected COVID-19 Infection. acr.org March 2020.
The Royal Australian and New Zealand College of Radiologist. COVID-19 Updates. ranzcr.com 2020
The Royal College of Radiologists. RCR position on the role of CT in patients suspected with COVID-19 infection Rcr.ac.uk 2020.
Canadian Association of Radiologists. Canadian Society of Thoracic Radiology and Canadian Association of Radiologists’ Statement on COVID -19. car.ca 2020.
Mossa-Basha M, Meltzer CC, Kim DC, Tuite MJ, Pallav Kolli K, Tan BS (2020) Radiology department preparedness for COVID-19: radiology scientific expert panel. Radiology. https://doi.org/10.1148/radiol.2020200988
Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ (2020) Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. https://doi.org/10.1148/radiol.2020200490
Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, Cui J, Xu W, Yang Y, Fayad ZA, Jacobi A, Li K, Li S, Shan H (2020) CT Imaging features of 2019 Novel coronavirus (2019-nCoV). Radiology 295(1):202–207
Wang Z, Gu S, Leader JK, Kundu S, Tedrow JR, Sciurba FC, Gur D, Siegfried JM, Pu J (2013) Optimal threshold in CT quantification of emphysema. Eur Radiol 23(4):975–984
Gattinoni L, Caironi P, Cressoni M et al (2020) Lung recruitment in patients with the acute respiratory distress syndrome. N Engl J Med 354(17):1775–1786
Ohkubo H, Kanemitsu Y, Uemura T, Takakuwa O, Takemura M, Maeno K, Ito Y, Oguri T, Kazawa N, Mikami R, Niimi A (2016) Normal lung quantification in usual interstitial pneumonia pattern: the impact of threshold-based volumetric CT analysis for the staging of idiopathic pulmonary fibrosis. PLoS ONE 11(3):e0152505
Belfiore MP, Urraro F, Grassi R, Giacobbe G, Patelli G, Cappabianca S, Reginelli A (2020) Artificial intelligence to codify lung CT in Covid-19 patients. Radiol Med. https://doi.org/10.1007/s11547-020-01195-x
Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C (2020) Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. https://doi.org/10.1148/radiol.2020200370
Albarello F, Pianura E, Di Stefano F, Cristofaro M, Petrone A, Marchioni L, Palazzolo C, Schininà V, Nicastri E, Petrosillo N, Campioni P, Eskild P, Zumla A, Ippolito G (2020) COVID 19 INMI Study Group. 2019-novel Coronavirus severe adult respiratory distress syndrome in two cases in Italy: an uncommon radiological presentation. Int J Infect Dis 93:192–197
Grassi R, Fusco R, Belfiore MP, Montanelli A, Patelli G, Urraro F, Petrillo A, Granata V, Sacco P, Mazzei MA, Feragalli B, Reginelli A, Cappabianca S (2020) Coronavirus disease 2019 (COVID-19) in Italy: features on chest computed tomography using a structured report system. Sci Rep. https://doi.org/10.21203/rs.3.rs-24312/v1
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan China. JAMA. https://doi.org/10.1001/jama.2020.1585
Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395(10223):514–523
Zhiliang Hu, Song Ci, Chuanjun Xu, Jin G, Chen Y, Xin Xu, Ma H, Chen W, Lin Y, Zheng Y, Wang J, Zhibin Hu, Yi Y, Shen H (2020) Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. https://doi.org/10.1007/s11427-020-1661-4
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS; China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus. https://doi.org/10.1056/NEJMoa2002032
Caruso D, Zerunian M, Polici M, Pucciarelli F, Polidori T, Rucci C, Guido G, Bracci B, de Dominicis C, Laghi A (2020) Chest CT features of COVID-19 in Rome. Italy Radiol 3:201237
Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K, Liu D, Wang G, Xu Q, Fang X, Zhang S, Xia J, Xia J (2020) Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology 296(2):2200905
Tárnok A (2020) Machine Learning, COVID-19 (2019-nCoV), and multi-OMICS. Cytom A 97(3):215–216
Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng C, Xu B (2020) A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv 2020.02.14.20023028.
Gozes O, Frid-Adar M, Greenspan H, Browning P, Zhang H, Ji W, Bernheim A, Siegel E (2020) Rapid AI Development Cycle for the Coronavirus (COVID-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring using Deep Learning CT Image Analysis. arXiv:2003.05037
van Ginneken B, Armato SG III, de Hoop B, Van de Amelsvoort-van VS, Duindam T, Niemeijer M, Murphy K, Schilham A, Retico A, Fantacci ME, Camarlinghi N, Bagagli F, Gori I, Hara T, Fujita H, Gargano G, Bellotti R, Tangaro S, Bolaños L, De Carlo F, Cerello P, Cristian Cheran S, Lopez TE, Prokop M (2010) Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: The ANODE09 study. Med Image Anal 14(6):707–722
Nardone V, Reginelli A, Guida C, Belfiore MP, Biondi M, Mormile M, Banci Buonamici F, Di Giorgio E, Spadafora M, Tini P, Grassi R, Pirtoli L, Correale P, Cappabianca S, Grassi R (2020) Delta-radiomics increases multicentre reproducibility: a phantom study. Med Oncol 37(5):38
Reginelli A, Belfiore MP, Monti R, Cozzolino I, Costa M, Vicidomini G, Grassi R, Morgillo F, Urraro F, Nardone V, Cappabianca S (2020) The Texture analysis as a predictive method in the assessment of the cytological specimen of CT guided FNAC of the lung cancer. Med Oncol. https://doi.org/10.1007/s12032-020-01375-9
Neri E, Coppola F, Miele V, Bibbolino C, Grassi R (2020) Artificial intelligence: Who is responsible for the diagnosis? Radiol Med 125(6):517–521
Bellotti R, De Carlo F, Gargano G, Tangaro S, Cascio D, Catanzariti E, Cerello P, Cheran SC, Delogu P, De Mitri I, Fulcheri C, Grosso D, Retico A, Squarcia S, Tommasi E, Golosio B (2007) A CAD system for nodule detection in low-dose lung CTs based on region growing and a new active contour model. Med Phys 34(12):4901–4910
Lachmann B (1992) Open up the lung and keep the lung open. Intens Care Med 18:319–321
Papadakos PJ, Lachmann B (2002) The open lung concept of alveolar recruitment can improve outcome in respiratory failure and ARDS. Mt Sinai J Med 69:73–77
Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, Schluger NW, Volpi A, Yim JJ, Martin IBK, Anderson DJ, Kong C, Altes T, Bush A, Desai SR, Goldin J, Goo JM, Humbert M, Inoue Y, Kauczor HU, Luo F, Mazzone PJ, Prokop M, Remy-Jardin M, Richeldi L, Schaefer-Prokop CM, Tomiyama N, Wells AU, Leung AN (2020) The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the fleischner society. Radiology 7:201365
Sverzellati N, Odone A, Silva M, Polverosi R, Florio C, Cardinale L, Cortese G, Addonisio G, Zompatori M, Dalpiaz G, Piciucchi S, Larici AR (2018) Italian structured report on fibrosing lung disease consort. structured reporting for fibrosing lung disease: a model shared by radiologist and pulmonologist. Radiol Med 123(4):245–253
Mohamed Hoesein FA, de Hoop B, Zanen P, Gietema H, Kruitwagen CL, van Ginneken B, Isgum I, Mol C, van Klaveren RJ, Dijkstra AE, Groen HJ, Boezen HM, Postma DS, Prokop M, Lammers JW (2011) CT-quantified emphysema in male heavy smokers: association with lung function decline. Thorax 66(9):782–787
Maldonado F, Moua T, Rajagopalan S, Karwoski RA, Raghunath S, Decker PA, Hartman TE, Bartholmai BJ, Robb RA, Ryu JH (2014) Automated quantification of radiological patterns predicts survival in idiopathic pulmonary fibrosis. Eur Respir J 43(1):204–212
Grassi R, Cappabianca S, Urraro F, Feragalli B, Montanelli A, Patelli G, Granata V, Giacobbe G, Russo GM, Grillo A, De Lisio A, Paura C, Clemente A, Gagliardi G, Magliocchetti S, Cozzi D, Fusco R, Belfiore MP, Grassi R, Miele V (2020) Chest CT computerized aided quantification of PNEUMONIA lesions in COVID-19 infection: a comparison among three commercial software. Int J Environ Res Public Health 17(18):E6914
Funding
No funding.
Author information
Authors and Affiliations
Contributions
All authors equally contributed to the investigations and to the writing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Human and animal Rights
“Bergamo Est” Institutional Review Institute (IRB) approved the study and renounced the written informed consent for this retrospective study, considering the ongoing epidemic emergency, which assessed the unidentified data and did not involve risk potential for patients.
Informed consent
Consent for publication was received.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Grassi, R., Belfiore, M., Montanelli, A. et al. COVID-19 pneumonia: computer-aided quantification of healthy lung parenchyma, emphysema, ground glass and consolidation on chest computed tomography (CT). Radiol med 126, 553–560 (2021). https://doi.org/10.1007/s11547-020-01305-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11547-020-01305-9