Abstract
Infection of the central nervous system with human immunodeficiency virus type 1 (HIV-1) can produce morphological changes in the neocortical synaptodendritic arbor that are correlated with neurocognitive impairment. To determine whether HIV-1 infection influences the protein composition of human synapses, a proteomic study of isolated nerve endings was undertaken. Synaptosomes from frontal neocortex were isolated using isopyknic centrifugation from 19 human brain specimens. Purity and enrichment were assessed by measuring pre- and postsynaptic protein markers. Two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption ionization time-of-flight mass spectrometry was used to screen for proteins differentially expressed in HIV/AIDS. The concentrations of 31 candidate protein spots were potentially abnormal in HIV-infected decedents with HIV encephalitis and/or increased expression of immunoproteasome subunits. Immunoblots showed that the concentration of some of them was related to HIV-1 infection of the brain and immunoproteasome (IPS) induction. Synapsin 1b and stathmin were inversely related to brain HIV-1 load; 14-3-3ζ and 14-4-4ε proteins were higher in subjects with HIV-1 loads. Perturbed synaptosome proteins were linked with IPS subunit composition, and 14-3-3ζ was histologically colocalized with IPS subunits in stained neocortical neurons. Proteomics illustrates that certain human proteins within the synaptic compartment are involved with changes in the synaptodendritic arbor and neurocognitive impairment in HIV-1-infected people.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infection with the human immunodeficiency virus type 1 (HIV-1) produces HIV-associated neurocognitive impairment (HAND) in a high proportion of subjects (Sacktor et al. 2001). Prior to the era of highly active antiretroviral therapy (HAART) in the USA, the prevalence of severe impairment was about 20%. After HAART, the severity of HAND was decreased very substantially, but its prevalence did not decrease. Fewer than 5% of HAART-treated clinical cohorts have outright dementia (McArthur 2004). The presumptive neuropathological substrate in some, but not all people with severe HAND is HIV encephalitis (HIVE; Glass et al. 1995; Wiley and Achim 1994). In HIVE, there is increased replication of HIV-1 in CNS glia, but not neurons, which leads to inflammation and morphological changes in neural elements (Kaul et al. 2001), including synapses, dendrites (Masliah et al. 1997), and axons (Giometto et al. 1997). Changes in the synaptodendritic arbor are potentially important because they are correlated significantly with HAND (Masliah et al. 1997). It is not clear whether abnormal neocortical synapses in HIVE represent irreversible neurodegeneration or instead reflect reversible physiological plasticity. Physiological plasticity of synapses and the concentration of synaptic proteins are both influenced locally in the synaptic compartment by the ubiquitin–proteasome system (UPS). The UPS is a main route of synaptic protein degradation, and it plays a central role in regulating the concentration of synaptic proteins, which influences synaptic plasticity and the minute-to-minute regulation of synaptic physiology (Bingol and Schuman 2005; Patrick 2006; Yi and Ehlers 2005). Disturbances of the UPS influence the synaptic protein economy and lead to morphological changes in the synaptodendritic compartment and physiological changes in synaptic transmission (Hegde 2004). For example, inhibition of UPS enzyme active sites (Fonseca et al. 2006) or exchanging UPS active sites with immunoproteasome (IPS) subunits (Gavilan et al. 2009; Maher et al. 2006) both produce an immediate disturbance of synaptic long term depression. In support of a potential role of HIV-1 on the UPS, it has been demonstrated histochemically and neurochemically that subjects with HAND accumulate ubiquitinylated protein conjugates in association with decreased synaptic protein (Gelman and Schuenke 2004). To further elucidate biochemical changes of proteins in the synaptic compartment in HIV/AIDS, we isolated nerve endings from brain neocortex (cerebrocortical synaptosomes) and performed a proteomic screening of synaptic proteins. The concentrations of certain synaptic proteins in the isolated nerve endings were related to replicating HIV-1 in the brain and induction of IPS subunit expression. These new observations provide increased support for the hypothesis that HIV-1 infection perturbs the protein economy of the synaptic compartment by modulation of the UPS.
Materials and methods
Subjects
Nineteen brain specimens from the National NeuroAIDS Tissue Consortium collection were used (Morgello et al. 2001). All subjects except for three uninfected normal control decedents had neuropsychological evaluations prior to death and underwent a complete autopsy and banking of the brain. All had natural deaths. Subjects and next-of-kin gave informed consent in accordance with statutes in the State of Texas in the USA. Protection of human subjects was regulated by the institutional review board at the University of Texas Medical Branch at Galveston. Neurocognitive performance was assessed in the HIV-1-infected subjects using a battery of tests (Morgello et al. 2001). The subjects were consolidated for confirmation studies using Western blotting as follows: normal brain not infected with HIV (n = 3), HIV-infected with low brain HIV-1 and low IPS concentration (n = 8), and HIV-infected with high HIV-1 and high IPS (n = 8). In the group with high HIV-1 and IPS, four of the subjects had HIVE and four did not. Concentrations of HIV-1 RNA in brain cortex in the three respective groups (log10 copies per gram of wet weight) were 0 ± 0, 2.77 ± 1.50, and 4.81 ± 0.48 (t test: p = 0.0000055, low versus high). The values for the IPS subunit LMP7 concentration in the three groups were 1.00 ± 0.79, 2.02 ± 1.02, and 4.03 ± 2.15 (t test: p = 0.0315, low versus high).
In the screening stage, these 19 samples were arranged into seven groups of pooled specimens to undertake two-dimensional polyacrylamide gel electrophoresis (2D-PAGE). These multiple groupings were designed to insure that we would account for potential influences of CNS impairment (HAND), IPS induction, and HIVE in at least one independent comparison. All seven of the synaptosome pools were produced with equal contributions of protein from its component samples as follows: pool A, normal HIV-negative subjects (n = 3); pool B, HIV-positive subjects without HAND (n = 3); pool C, HIV-positive subjects with HAND but without HIVE (n = 3); pool D, HIV-positive subjects with HAND and HIVE (n = 3); pool E, HIV-positive with low IPS expression (n = 3); pool F, HIV-positive with high IPS and no HIVE (n = 3); and pool G, HIV-infected with high IPS and HIVE (n = 4). To cover all of the desired comparisons in the screening stage, a few synaptosome isolates were used in more than one pool. Seven gels for pools A through G were each analyzed in triplicate on 2D-PAGE. Computerized analysis of digital images of the 21 gels was performed to compare spot intensities between groups, which are as follows: A versus B, B versus C, B versus D, C versus D, E versus F, E versus G, and F versus G. After these preliminary comparisons were done and candidate spots were identified (see below), synaptosome isolates from each of the 19 subjects were then collapsed into three main groups described above to undertake confirmatory studies using Western blotting.
Synaptosome isolation
About 500 mg of dorsolateral prefrontal cortex from Brodmann area 9 was dissected free while frozen. Synaptosomes were isolated at 4°C by differential centrifugation in a discontinuous sucrose gradient using a standard procedure (Dodd et al. 1981; Eshleman et al. 2001; Gray and Whittaker 1962; Mash et al. 2002; Wood et al. 1996). Specimens were thawed briefly and added to 5 mL of ice cold buffered sucrose (0.32 M sucrose, 5 mM HEPES, 25 μL protease inhibitor cocktail (Sigma Aldrich), and 50 μL phosphatase inhibitor cocktail (EMD Chemicals, Inc., Gibbstown, NJ, USA), pH 7.4). They were homogenized with eight strokes in a Potter–Elvehjem tissue grinder with 0.1 to 0.5 mm tolerance (Fisher Scientific), with intermittent immersion in an ice bath for 30 s between strokes. Homogenates were centrifuged at 1,000×g for 10 min at 4°C to yield a crude pellet (P1a) and supernatant (S1a). P1a was washed and centrifuged as above, yielding a second pellet (P1b) and supernatant (S1b). The supernatants were combined into S1, brought to a volume of 10 mL, and centrifuged at 10,000×g for 20 min at 4°C to produce the P2 pellet. The P2 pellet was resuspended in 2.5 mL buffered sucrose and layered over a discontinuous gradient composed of 2.5 mL each of buffered sucrose with concentrations of 0.8, 1.0, and 1.2 M from top to bottom. P2 was centrifuged at 150,000×g for 2 h at 4°C. The P3 synaptosome fraction was collected from the interface of the 1.0- and 1.2-M layers. P3 was washed in 0.32 M buffered sucrose at 150,000×g for 30 min at 4°C. The P3 pellet was collected and stored frozen until use. The relative enrichment of synaptic protein markers and depletion of glial markers in these synaptosome isolates was compared to the starting homogenate using Western blotting of pre- and postsynaptic protein markers, as listed in Fig. 1.
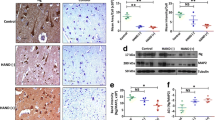
Immunoblots show that human cerebrocortical synaptosome preparations from frontal neocortex specimens of eight human subjects are enriched with synaptic marker proteins. a The immunoproteasome subunit LMP7 was increased in synaptosomes from the HIV-positive subjects. Pre- (b) and postsynaptic (c) marker proteins were enriched in synaptosomes by up to 7-fold when compared to unfractionated homogenates. d Marker proteins for non-neural cells, including mononuclear phagocytes (CD68) and astrocytes (GFAP) were sharply depleted in these synaptosome isolates. See Table 1 for added description of the proteins
Proteomic screening
Synaptosomes were pooled into the seven groups, and then 2D-PAGE was performed in triplicate in the proteomics core laboratory at the University of Texas Medical Branch (http://www.utmb.edu/brf/). Isoelectric focusing was done in the first dimension and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in the second dimension. Gels were stained with SYPRO Red and photographed, and images were analyzed by Nonlinear Dynamics Progenesis SameSpots software (Nonlinear USA, Inc, Durham, NC, USA). Spots with at least a 2-fold difference in averaged optical density were considered further. Spots that had low intensity, were part of a streak, or were not present in at least two gels were eliminated. The gel with the highest spot intensity was selected for manual excision for evaluation by mass spectrometry using a large-bore pipette tip.
Mass spectrometry
Gel spot samples were cut into 1-mm-size pieces or smaller and placed into separate 0.5 mL polypropylene tubes; 100 μL of 50 mm ammonium bicarbonate buffer was added to each tube, and the samples were incubated at 37°C for 30 min. The buffer was removed, and 100 μL of water was then added to each tube. The samples were incubated again at 37°C for 30 min, water was removed, and 100 μL of acetonitrile was added to dehydrate the gel pieces. After 5 min, the acetonitrile was removed, and 100 μL of acetonitrile was again added and removed. Excess solvent was removed in a Speedvac for 45 min and then 2 mL of 25 mM ammonium bicarbonate at pH 8.0 was added to a 20-μg vial of lyophilized trypsin (Promega Corporation, Madison, WI, USA). The trypsin solution was added to the sample, and they were digested at 37°C for 6 h. One microliter of sample solution was spotted directly onto a matrix-assisted laser desorption ionization (MALDI) target plate and allowed to dry. One microliter of alpha-cyano-4-hydroxycinnamic acid (Sigma Aldrich) matrix solution (50:50 acetonitrile/water at 5 mg/mL) was then applied on the sample spot and allowed to dry. The dried MALDI spot was blown with compressed air (Decon Laboratories, Inc, King of Prussia, PA, USA) before mass spectrometry.
MALDI time-of-flight mass spectrometry (TOF-MS) was used to obtain protein identification. Data were acquired with an Applied Biosystems 4800 MALDI TOF/TOF Proteomics Analyzer (Applied Biosystems, Foster City, CA, USA). Applied Biosystems software package included 4000 Series Explorer (v. 3.6 RC1) with Oracle Database Schema Version (v. 3.19.0), Data Version (3.80.0) to acquire both MS and MS/MS spectral data. The instrument was operated in positive ion reflectron mode, mass range was 850–3,000 Da, and the focus mass was set at 1,700 Da. For MS data, 1,000–2,000 laser shots were acquired and averaged from each sample spot. Automatic external calibration was performed using a peptide mixture with reference masses 904.468, 1,296.685, 1,570.677, and 2,465.199.
MALDI MS/MS was performed on several (five to 10) abundant ions from each sample spot. A 1-kV positive ion, MS/MS method was used to acquire data under postsource decay (PSD) conditions. The instrument precursor selection window was ±3 Da. For MS/MS data, 2,000 laser shots were acquired and averaged from each sample spot. Automatic external calibration was performed using reference fragment masses 175.120, 480.257, 684.347, 1,056.475, and 1,441.635 (from precursor mass 1,570.700). Applied Biosystems GPS ExplorerTM (v. 3.6) software was used in conjunction with MASCOT to search the respective protein database using both MS and MS/MS spectral data for protein identification. Protein match probabilities were determined using expectation values and/or MASCOT protein scores. MS peak filtering included the following parameters: mass range 800 to 3,000 Da, minimum S/N filter = 10, mass exclusion list tolerance = 0.5 Da, and mass exclusion list (for some trypsin and keratin-containing compounds) included masses 842.51, 870.45, 1,045.56, 1,179.60, 1,277.71, 1,475.79, and 2,211.1. For MS/MS peak filtering, the minimum S/N filter = 10.
For protein identification, the human taxonomy was searched in the database. Other conditions used included selection of trypsin as the digesting enzyme, the maximum missed cleavages was =1, a fixed modification was carbamidomethyl (C) for 2-D gel analyses only, a variable modifications was oxidation (M), precursor tolerance was set at 0.2 Da, MS/MS fragment tolerance was set at 0.3 Da, mass = monoisotopic, and peptide charges were only considered as +1. The significance of a protein match, based on both the peptide mass fingerprint in the first MS and the MS/MS data from several precursor ions, was based on expectation values; each protein match was accompanied by an expectation value. The expectation value was the number of matches with equal or better scores that were expected to occur by chance alone. The default significance threshold was p < 0.05 (an expectation value of 0.05 is considered to be on this threshold). A more stringent threshold of 10−3 for protein identification was used; the lower the expectation value, the more significant the score.
Western blotting
Total protein (10−30 μg) in 15 μL volume of synaptosome or earlier fractions was added to 15 μL of 2× Laemmli sample buffer (Bio-Rad Laboratories) with 5% beta-mercaptoethanol in 1.5 mL microcentrifuge tubes and boiled for 5 min. Samples were then loaded into Criterion Precast 18-well 5%, 15%, or 4–20% gradient Tris–HCL gels (Bio-Rad Laboratories) for SDS-PAGE to run for approximately 1 h at 180 V. Proteins were transferred to polyvinylidene fluoride membrane (Amersham Biosciences, Piscataway, NJ, USA) for 3 h at 60 V in 4°C. Membranes were blocked with 5% nonfat dry milk in TBST (0.05 M Trizma–HCl, 0.15 M NaCl, and 0.1% Tween 20) for 1 h and then incubated overnight with primary antibody (Table 1) diluted in TBST according to the antibody supplier data sheet. Membranes were then washed three times for 5 min with TBST and incubated with the appropriate anti-rabbit or anti-mouse secondary antibody diluted 1:5,000 for 1 h (Amersham Biosciences). After washing, Enhanced Chemiluminescence Detection Reagent (Amersham Biosciences) was applied for 2 min, and the membrane was exposed to Kodak BioMAX XAR film (Rochester, NY, USA) for 30 s to 3 min, depending upon signal intensity. The film was developed and digitized by computer scanner, and band density was quantified using One-Dscan (BD Biosciences Bioimaging, Rockville, MD, USA).
Immunofluorescence microscopy
Paraffin-embedded sections of dorsolateral prefrontal cortex from subjects with HIVE and controls were used. Antigen retrieval was performed in sodium citrate buffer using a preheated vegetable steamer for 20 min. Sections were cooled and then blocked with Image-iT FX signal enhancer (Invitrogen Molecular Probes, Eugene, OR, USA) for 30 min, followed by blocking with 5% bovine serum albumin plus 5% normal goat serum in TBST for 1 h. Primary antibodies (Table 1) were diluted in blocking solution and applied overnight at 4°C. Slides were incubated for 1 h with the appropriate Alexa-Fluor fluorochrome-conjugated secondary antibody (Invitrogen Molecular Probes) diluted in blocking solution. To block autofluorescence, sections were incubated with Sudan Black B (Sigma Aldrich) solution (1% Sudan Black B in 70% ethanol) for 10 min. After washing, coverslips were mounted using Slow Fade Gold with DAPI (Invitrogen Molecular Probes) mounting medium. Laser confocal microscopy was performed with a Zeiss LSM 510 UV META laser scanning confocal microscope consisting of an Axiovert 200 M Inverted Microscope equipped with an oil-immersion ×100 resolution objective for fluorescence, Ar, dual HeNe, and UV lasers and fluorescence filters set for DAPI, FITC, TRITC, and far red, a scanning module with visible and UV acousto optical tunable filters, two independent fluorescence channels (2 PMTs), and a 32-PMT array (Carl Zeiss MicroImaging, Inc, Thornwood, NY, USA).
HIV-1 RNA concentration
Detection of the HIV-1 envelope protein in mRNA extracts from brain tissue was performed using a modification of a single copy procedure (Palmer et al. 2003). HIV RNA from frontal neocortex was extracted using RNeasy Lipid Tissue Mini Kit (Qiagen, Valencia, CA, USA). One microgram of brain RNA and 1 μmol/L of anti-sense primer 84R were used in 20 μL reactions (iScript cDNA Synthesis Kit, Bio-Rad, Hercules, CA, USA). Four microliters of cDNA was used for a 25 μL real-time PCR using JumpStart Taq ReadyMix (Sigma, Saint Louis, MO, USA) and SmartCycler (Cepheid, Sunnyvale, CA, USA). Results were standardized against a known brain secondary standard and expressed according to wet weights. Cerebrospinal fluid (CSF) and blood plasma HIV-1 RNA concentration (copies per milliliter) was measured using the Amplicor HIV-1 Monitor® Test (Roche Diagnositics Inc., Indianapolis, IN, USA).
Statistics
Data were compiled and analyzed using Microsoft Excel 2003 (Microsoft Corporation, Redmond, Washington, DC, USA) and GraphPad InStat Version 3.06 (GraphPad Software, Inc., La Jolla, CA, USA). Student’s t test was performed in Microsoft Excel 2003 to determine differences between two groups. Comparisons between three or more groups were performed by analysis of variance (ANOVA) with posttest analyses in GraphPad Instat. Data were tested for Gaussian distribution by the method of Kolmogorov and Smirnov. If the data were considered normal, one-way ANOVA was performed, followed by the Tukey–Kramer multiple comparisons test. Data that did not pass the normality test were analyzed by the Kruskal–Wallis test (non-parametric ANOVA) followed by Dunn's multiple comparisons test. Analysis of correlation and regression was performed in Microsoft Excel. Graphs were produced in Microsoft Excel 2003. Where applicable, error bars represent an approximation of the standard error of the mean by the 95% confidence intervals.
Results
Protein composition of human synaptosome isolates
All human brain neocortical specimens were frozen at −80°C prior to isolation of synaptosomes, except in preliminary experiments to compare protein yields using freshly obtained brain specimens and/or cryoprotected and rapidly thawed brain specimens from the same individual. The distribution and the enrichment of synaptic protein markers in preparations from the frozen samples was identical to freshly prepared unfrozen and cryoprotected frozen specimens from the same decedent (not shown). Postsynaptic protein markers were enriched about 6-fold in synaptosome prepared from fresh tissue, frozen tissue, and cryoprotected tissue; presynaptic markers were less markedly enriched (Fig. 1). Glial cell markers, including glial fibrillary acidic protein, were sharply depleted in the preparations. These results are generally equivalent to those obtained using a preparation from a fresh or frozen rodent brain (Dodd et al. 1981; Eshleman et al. 2001; Gray and Whittaker 1962; Mash et al. 2002; Wood et al. 1996). Osmotic lysis of human cerebrocortical synaptosomes produced further protein enrichment (not illustrated), but we did not employ osmotic lysis in order to prevent the release of membrane-bound proteasome components.
Preliminary spot lists from the screening gels
The yield of synaptosome protein from brain homogenates and the time and effort required to perform the isolation served to limit the number of subjects that could be analyzed. The proteomic analysis was designed to compare seven synaptosome pools that were produced from the isolates of 19 subjects. From the seven screening pools, seven comparisons were done; a total of 139 spots with at least a 2-fold difference in optical density were identified. The preliminary “spot list” was reduced to 68 spots by eliminating manually those spots that were too low in intensity or were not present in at least two of the three gels. For the remaining spots, the gel with the highest spot intensity was selected, and the area was removed using a large-bore pipette tip. Each spot was analyzed by mass spectrometry. After MS analysis, 31 candidate proteins were identified (Table 2).
Immunoblotting
Nine promising protein candidates identified by mass spectrometry were evaluated in individual synaptosome extracts by Western blotting. Three proteins were significantly different in one or more of the HIV-infected groups; another protein was changed marginally (Fig. 2a). Synapsin 1b concentration was sharply decreased in the HIV-positive subjects with high brain HIV-1 and LMP7. 14-3-3ζ concentration was increased by 75% in the subjects with high brain HIV-1 and LMP7 (p < 0.01). Synaptosome 14-3-3ε was increased about 2-fold in the HIV-positive subjects with high HIV-1 and LMP7, as compared to those with low HIV-1 (p < 0.05). When the protein concentrations were compared to HIV-1 concentrations in brain, CSF, and blood, Pearson correlation coefficients were positive and significant for brain and CSF HIV-1 loads (Table 3). Stathmin was marginally decreased in the high HIV-1 group (Fig. 2c) and was significantly correlated with brain HIV-1 (Table 3). Immunoblots confirmed that the IPS subunit LMP7 was increased 3-fold in synaptosome and whole cell preparations both (Figs. 1 and 2a). Concentrations of six other proteins on the original list of 31 candidates in Table 2 were not significantly altered in the immunoblots (Fig. 2c).
Immunoblots of synaptosomes prepared from frontal neocortex from all 19 human subjects. Comparison groups are according to status of HIV-1 infection, neocortical HIV-1 RNA load (low and high), and neocortical immunoproteasome subunit expression (low and high). a Subjects with high HIV-1 loads had significantly more synaptosome LMP7 as compared to the other groups. b Synapsin 1b concentration was decreased in subjects with high HIV-1 load and abundant LMP7; 14-3-3 zeta and 14-3-3 epsilon were increased with high HIV-1 and LMP7. c The decreased stathmin in subjects with high HIV-1 and LMP7 was marginal (p < 0.1). Concentrations of HIV-1 RNA in brain cortex in the three respective groups (log10 copies per gram of wet weight) were 0 ± 0, 2.77 ± 1.50, and 4.81 ± 0.48 (t test: p = 0.0000055, low versus high). The values for the IPS subunit LMP7 concentration in the three groups were 1.00 ± 0.79, 2.02 ± 1.02, and 4.03 ± 2.15 in relative density units (t test: p = 0.0315, low versus high). Six other candidate protein species tested were not different statistically. IPS neocortical immunoproteasome concentration. Mean ± one standard deviation. *p < 0.05; **p < 0.01
Morphological localization with immunoproteasomes
To determine whether the altered synaptosomal proteins in HIV/AIDS are histologically related to IPS subunits, we performed dual localization using laser confocal microscopy. Immunoreactivities of 14-3-3ζ and the IPS subunit LMP2 both were present in punctate neuronal and neuropil markings that were typical of neocortical synapses. Colocalization of 14-3-3ζ and LMP2 in these punctate synaptic deposits was often observed (Fig. 3). Colocalizing proteins were most obvious in the brains that contained a high HIV RNA concentration and HIVE. It was not possible to demonstrate a spatial relationship using synapsin 1b staining because its concentration was sharply depressed due to HIV-1 and difficult to detect histochemically (not illustrated).
Confocal immunofluorescence microscopy for 14-3-3ζ (red) and the immunoproteasome subunit marker LMP2 (green) in a subject with HIV encephalitis and high concentrations of these proteins in the synaptosome preparation. Punctate deposits of both proteins outline the distinct shape of a pyramidal neocortical neuron and surrounding neuropil. The perikaryon and neuropil both contain deposits of these proteins, which morphologically resemble synaptic densities. Arrows in the large bottom panel denote foci where the antigens are colocalized in the merged image. Attempts to localize synapsin 1 were not successful technically because its concentration was sharply decreased in HIV encephalitis. Scale bar = 10 μm
Discussion
This proteomic analysis focused on the synaptic compartment because HIVE produces histological changes in the synaptodendritic arbor (Masliah et al. 1997) and biochemical changes in synaptic proteins (Gelman and Schuenke 2004). Isolated nerve endings from humans infected with HIV-1 contained subtle and specific changes in their protein composition. The concentrations of three synaptic proteins were abnormal in brain fractions that were enriched in synaptic nerve endings. The concentration of the presynaptic protein synapsin 1b was sharply decreased in synaptosomes from subjects with a high HIV-1 and immunoproteasome concentration in brain neocortex. Stathmin had a statistically marginal decrease and was negatively correlated with brain HIV-1 load. The concentrations of 14-3-3ζ and 14-3-3ε were significantly greater in the high brain HIV-1 group. A wide range of pre- and postsynaptic marker proteins was not affected by HIV-1 infection (Fig. 1), which illustrates that a comprehensive decline of the entire synaptic protein repertoire did not occur.
We focused attention on the hypothesis that the multicatalytic proteinase known as the UPS might be correlative. The UPS operates locally in the synaptic compartment and exerts a strong and physiologically rapid influence on synaptic protein concentrations (Yi and Ehlers 2005). It is important, therefore, that the protein changes we observed were correlated with induction of immunoproteasome subunits (IPS) of the UPS (e.g., LMP7; Figs. 1 and 2a), which is an innate response to virus infection (Heink et al. 2005). Indeed, the synaptosome protein changes were related significantly to HIV-1 loads in brain tissue and cerebrospinal fluid (Table 3). One of the proteins was histologically colocalized with LMP7 in punctate neuronal structures morphologically consistent with synapses (Fig. 3). Using scarce specimens from human subjects, these correlative results suggest that the IPS may be involved with the local regulation of synaptic proteins in HIV-1 infection. In turn, synaptosome protein changes associated with IPS induction could lead to morphological changes in the synaptodendritic arbor, which is correlated with HIV-1-associated neurocognitive impairment (Masliah et al. 1997).
Synapsin 1b is a phosphoprotein localized primarily in presynaptic nerve terminals. It modulates the availability of synaptic vesicles bound to cytoskeletal proteins and controls neurotransmitter release in response to action potentials (Evergren et al. 2007). Knockout mice lacking synapsin genes have severe synaptic depression after repetitive high stimulation; the protein probably plays a key role in regulating the reserve pool of synaptic vesicles (Rosahl et al. 1995; Chi et al. 2001; Gitler et al. 2004; Sun et al. 2006). Its concentration has been noted to decrease in senile dementia of the Alzheimer type, although this could reflect a nonspecific loss of whole synapses instead of a specific change within viable nerve endings, as suggested for HIV/AIDS (Ho et al. 2001; Qin et al. 2004; Sze et al. 2000). The decrease of synapsin 1b in nerve endings associated with HIV-1 infection suggests that there is abnormal regulation of the reserve pool of presynaptic vesicles of brain neocortex, analogous to what happens in synapsin 1 knockout mice. Physiologically, knocking out synapsin 1 gene (Gitler et al. 2004), inhibiting the UPS (Fonseca et al. 2006), and increased LMP7 and IPS synthesis (Gavilan et al. 2009; Maher et al. 2006) all produce long-term synaptic depression (LTD). When synapsin 1 is downregulated in LTD, the effect is strictly dependent on normally functioning active sites of the UPS (Fioravante et al. 2008). Our results using human synaptosomes support the suggested relationship between decreased concentration of synapsin 1b and the UPS (i.e., the increased immunoproteasome active site, LMP7; Fig. 2a). The potential role of HIV-1 on posttranscriptional regulation of synaptosomal synapsin 1 by the UPS and its potential relationship to LTD are aspects that warrant further elucidation in the future. A degradation pathway for synapsin 1 via calcium-activated calpain proteases also might be involved in synapsin downregulation (Murrey et al. 2006).
The decrease of stathmin in synaptosomes was marginal statistically, yet worthy of notice because unlike synapsin, it can be localized in postsynaptic dendrites. Stathmin is a phosphoprotein that plays roles in synaptic physiology, axon aging, and maintenance. It regulates the balance between polymerization and depolymerization of microtubules, which is critical for elongation of dendritic arbors. Knockdown of stathmin increases microtubule stability and decreases dendritic elongation (Grenningloh et al. 2004). Stathmin knockout mice display a defect in maintaining axon stability during aging (Liedtke et al. 2002). The decrease of stathmin concentration in synaptosomes in HIV-1 infection could modify the synaptodendritic arbor (Masliah et al. 1997) and axon integrity (Giometto et al. 1997) via stabilization of microtubule assemblies and decreased turnover of microtubule proteins.
The 14-3-3 protein family is highly abundant in the brain. Its members modulate protein interactions and regulate many neuronal processes (Berg et al. 2003a; Kjarland et al. 2006), including synaptic plasticity (Dai and Murakami 2003), neurotransmitter synthesis (Ichimura et al. 1987; Kleppe et al. 2001), and synaptic ion channel function and localization (Bunney et al. 2002; Rajan et al. 2002; Suginta et al. 2001; Zhou et al. 1999). Given their widespread functions, it is difficult to predict the functional impact of having increased 14-3-3 proteins in HIV-1 infection. Its ion channel activation properties can modify long-term potentiation and LTD (Zhou et al. 1999), similar to what happens when synapsin 1 and stathmin are perturbed. The concentration of 14-3-3 proteins is linked to several neurodegenerative diseases, including HAND. Demented patients with spongiform encephalopathies have increased concentrations of 14-3-3 proteins in the CSF (Berg et al. 2003a; Kenney et al. 2000; Lemstra et al. 2000); these proteins accumulate neuropathologically in characteristic Prion Protein (PrP) plaques (Richard et al. 2003). 14-3-3 proteins localize within neurons that contain neurofibrillary tangles, a key neuropathological change in senile dementia (Hashiguchi et al. 2000; Umahara et al. 2004). They also are present in Lewy bodies, which are the pathological hallmark of Parkinson disease and Lewy body dementias (Berg et al. 2003b; Kawamoto et al. 2002). 14-3-3 proteins epsilon, gamma, and zeta isoforms (but not beta, eta, or tau) have been reported to be increased in the CSF of patients with HIV/AIDS, primarily those with HIV-associated dementia or cytomegalovirus infection (Miller et al. 2000; Wakabayashi et al. 2001). In macaques infected with simian immunodeficiency virus (SIV), 14-3-3 proteins in the CSF were increased primarily in animals with high SIV loads in brain (Helke et al. 2005). Rodents infected with a murine immunodeficiency virus had decreased 14-3-3ζ concentration in whole hippocampus (CSF not measured; Takahashi et al. 2007). Since 14-3-3ζ is involved in synaptic plasticity and neurotransmitter release, its anomalous concentrations in HIV-1-infected synaptosomes may reflect changes in the synaptodendritic arbor, which occur primarily with high HIV-1 loads in the brain (Masliah et al. 1997). Regulation of ion channels by 14-3-3 species can produce acquired neuronal channelopathies, which have been documented to occur in HAND (Gelman et al. 2004).
Summary
Proteomics using isolated nerve endings has revealed novel synaptic protein changes in HIV-1-infected human subjects. Synapsin 1b, 14-3-3ζ, 14-3-3ε, and stathmin concentrations were selectively perturbed and were correlated with brain HIV-1 load and altered expression of UPS subunits. The regulated proteins play key roles in neurotransmitter release, dendritic morphology, synaptic plasticity, and long-term synaptic depression. Their abnormal concentrations in the synaptic compartment could be linked to morphological changes in the synaptodendritic arbor and neuronal channelopathies in HAND. Physiological regulation of proteins by the UPS locally within the synaptic compartment is a well-documented pathway experimentally; these new results with human nerve endings support the suggestion that the UPS could be involved in HIV-associated changes in synaptic proteins. Alternate screening strategies could reveal still other abnormal protein species of the synaptic proteome due to HIV-1, as many proteins species were not within range of the chosen technique.
References
Berg D, Holzmann C, Riess O (2003a) 14-3-3 proteins in the nervous system. Nat Rev Neurosci 4:752–762
Berg D, Riess O, Bornemann A (2003b) Specification of 14-3-3 proteins in Lewy bodies. Ann Neurol 54:135
Bingol B, Schuman EM (2005) Synaptic protein degradation by the ubiquitin proteasome system. Curr Opin Neurobiol 15:536–541
Bunney TD, van den Wijngaard PW, de Boer AH (2002) 14-3-3 protein regulation of proton pumps and ion channels. Plant Mol Biol 50:1041–1051
Chi P, Greengard P, Ryan TA (2001) Synapsin dispersion and reclustering during synaptic activity. Nat Neurosci 4:1187–1193
Dai JG, Murakami K (2003) Constitutively and autonomously active protein kinase C associated with 14-3-3 zeta in the rodent brain. J Neurochem 84:23–34
Dodd PR, Hardy JA, Oakley AE, Edwardson JA, Perry EK, Delaunoy JP (1981) A rapid method for preparing synaptosomes: comparison, with alternative procedures. Brain Res 226:107–118
Eshleman AJ, Wolfrum K, Mash DC, Christensen K, Janowsky A (2001) Drug interactions with the dopamine transporter in cryopreserved human caudate. J Pharmacol Exp Ther 296:442–449
Evergren E, Benfenati F, Shupliakov O (2007) The synapsin cycle: a view from the synaptic endocytic zone. J Neurosci Res 85:2648–2656
Fioravante D, Liu R-Y, Byrne JH (2008) The ubiquitin–proteasome system is necessary for long-term synaptic depression in aplysia. J Neurosci 28:10245–10256
Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV (2006) A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52:239–245
Gavilan MP, Castano A, Torres M, Portavella M, Caballero C, Jimenez S, Garcia-Martinez A, Parrado J, Vitorica J, Ruano D (2009) Age-related increase in the immunoproteasome content in rat hippocampus: molecular and functional aspects. J Neurochem 108:260–272
Gelman BB, Schuenke K (2004) Brain aging in acquired immunodeficiency syndrome: increased ubiquitin-protein conjugate is correlated with decreased synaptic protein but not amyloid plaque accumulation. J Neurovirol 10:98–108
Gelman BB, Soukup VM, Keherly MJ, Schuenke KW, Holzer CE, Richey JL, Lahart C (2004) Acquired neuronal channelopathies in HIV-associated dementia. J Neuroimmunology 157:111–119
Giometto B, An SF, Groves M, Scaravilli T, Geddes JF, Miller R, Tavolato B, Beckett AA, Scaravilli F (1997) Accumulation of beta-amyloid precursor protein in HIV encephalitis: relationship with neuropsychological abnormalities. Ann Neurol 42:34–40
Gitler D, Takagishi Y, Feng J, Ren Y, Rodriguiz RM, Wetsel WC, Greengard P, Augustine GJ (2004) Different presynaptic roles of synapsins at excitatory and inhibitory synapses. J Neurosci 24:11368–11380
Glass JD, Fedor H, Wesselingh SL, McArthur JC (1995) Immunocytochemical quantitation of human immunodeficiency virus in the brain: correlations with dementia. Ann Neurol 38:755–762
Gray EG, Whittaker VP (1962) The isolation of nerve endings from brain: an electron-microscopic study of cell fragments derived by homogenization and centrifugation. J Anat 96:79–88
Grenningloh G, Soehrman S, Bondallaz P, Ruchti E, Cadas H (2004) Role of the microtubule destabilizing proteins SCG10 and stathmin in neuronal growth. J Neurobiol 58:60–69
Hashiguchi M, Sobue K, Paudel HK (2000) 14-3-3zeta is an effector of tau protein phosphorylation. J Biol Chem 275:25247–25254
Hegde AN (2004) Ubiquitin–proteasome-mediated local protein degradation and synaptic plasticity. Prog Neurobiol 73:311–357
Heink S, Ludwig D, Kloetzel PM, Kruger E (2005) IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci U S A 102:9241–9246
Helke KL, Queen SE, Tarwater PM, Turchan-Cholewo J, Nath A, Zink MC, Irani DN, Mankowski JL (2005) 14-3-3 protein in CSF: an early predictor of SIV CNS disease. J Neuropathol Exp Neurol 64:202–208
Ho L, Guo Y, Spielman L, Petrescu O, Haroutunian V, Purohit D, Czernik A, Yemul S, Aisen PS, Mohs R, Pasinetti GM (2001) Altered expression of a-type but not b-type synapsin isoform in the brain of patients at high risk for Alzheimer's disease assessed by DNA microarray technique. Neurosci Lett 298:191–194
Ichimura T, Isobe T, Okuyama T, Yamauchi T, Fujisawa H (1987) Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+, calmodulin-dependent protein kinase II. FEBS Lett 219:79–82
Kaul M, Garden GA, Lipton SA (2001) Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature 410:988–994
Kawamoto Y, Akiguchi I, Nakamura S, Honjyo Y, Shibasaki H, Budka H (2002) 14-3-3 proteins in Lewy bodies in Parkinson disease and diffuse Lewy body disease brains. J Neuropathol Exp Neurol 61:245–253
Kenney K, Brechtel C, Takahashi H, Kurohara K, Anderson P, Gibbs CJ Jr (2000) An enzyme-linked immunosorbent assay to quantify 14-3-3 proteins in the cerebrospinal fluid of suspected Creutzfeldt–Jakob disease patients. Ann Neurol 48:395–398
Kjarland E, Keen TJ, Kleppe R (2006) Does isoform diversity explain functional differences in the 14-3-3 protein family? Curr Pharm Biotechnol 7:217–223
Kleppe R, Toska K, Haavik J (2001) Interaction of phosphorylated tyrosine hydroxylase with 14-3-3 proteins: evidence for a phosphoserine 40-dependent association. J Neurochem 77:1097–1107
Lemstra AW, van Meegen MT, Vreyling JP, Meijerink PH, Jansen GH, Bulk S, Baas F, van Gool WA (2000) 14-3-3 testing in diagnosing Creutzfeldt-Jakob disease: a prospective study in 112 patients. Neurology 55:514–516
Liedtke W, Leman EE, Fyffe RE, Raine CS, Schubart UK (2002) Stathmin-deficient mice develop an age-dependent axonopathy of the central and peripheral nervous systems. Am J Pathol 160:469–480
Maher FO, Clarke RM, Kelly A, Nally RE, Lynch MA (2006) Interaction between interferon gamma and insulin-like growth factor-1 in hippocampus impacts on the ability of rats to sustain long-term potentiation. J Neurochem 96:1560–1571
Mash DC, Pablo J, Ouyang Q, Hearn WL, Izenwasser S (2002) Dopamine transport function is elevated in cocaine users. J Neurochem 81:292–300
Masliah E, Heaton RK, Marcotte TD, Ellis RJ, Wiley CA, Mallory M, Achim CL, McCutchan JA, Nelson JA, Atkinson JH, Grant I (1997) Dendritic injury is a pathological substrate for human immunodeficiency virus-related cognitive disorders. HNRC Group. The HIV Neurobehavioral Research Center. Ann Neurol 42:963–972
McArthur JC (2004) HIV dementia: an evolving disease. J Neuroimmunol 157:3–10
Miller RF, Green AJ, Giovannoni G, Thompson EJ (2000) Detection of 14-3-3 brain protein in cerebrospinal fluid of HIV infected patients. Sex Transm Infect 76:408
Morgello S, Gelman BB, Kozlowski PB, Vinters HV, Masliah E, Cornford M, Cavert W, Marra C, Grant I, Singer EJ (2001) The National NeuroAIDS Tissue Consortium: a new paradigm in brain banking with an emphasis on infectious disease. Neuropathol Appl Neurobiol 27:326–335
Murrey HE, Gama CI, Kalovidouris SA, Luo WI, Driggers EM, Porton B, Hsieh-Wilson LC (2006) Protein fucosylation regulates synapsin Ia/Ib expression and neuronal morphology in primary hippocampal neurons. Proc Natl Acad Sci U S A 103:21–26
Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM (2003) New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 41:4531–4536
Patrick GN (2006) Synapse formation and plasticity: recent insights from the perspective of the ubiquitin proteasome system. Curr Opin Neurobiol 16:90–94
Qin S, Hu XY, Xu H, Zhou JN (2004) Regional alteration of synapsin I in the hippocampal formation of Alzheimer's disease patients. Acta Neuropathol 107:209–215
Rajan S, Preisig-Muller R, Wischmeyer E, Nehring R, Hanley PJ, Renigunta V, Musset B, Schlichthorl G, Derst C, Karschin A, Daut J (2002) Interaction with 14-3-3 proteins promotes functional expression of the potassium channels TASK-1 and TASK-3. J Physiol 545:13–26
Richard M, Biacabe AG, Streichenberger N, Ironside JW, Mohr M, Kopp N, Perret-Liaudet A (2003) Immunohistochemical localization of 14.3.3 zeta protein in amyloid plaques in human spongiform encephalopathies. Acta Neuropathol 105:296–302
Rosahl TW, Spillane D, Missler M, Herz J, Selig DK, Wolff JR, Hammer RE, Malenka RC, Sudhof TC (1995) Essential functions of synapsins I and II in synaptic vesicle regulation. Nature 375:488–493
Sacktor N, Lyles RH, Skolasky R, Kleeberger C, Selnes OA, Miller EN, Becker JT, Cohen B, McArthur JC (2001) HIV-associated neurologic disease incidence changes: Multicenter AIDS Cohort Study, 1990–1998. Neurology 56:257–260
Suginta W, Karoulias N, Aitken A, Ashley RH (2001) Chloride intracellular channel protein CLIC4 (p64H1) binds directly to brain dynamin I in a complex containing actin, tubulin and 14-3-3 isoforms. Biochem J 359:55–64
Sun J, Bronk P, Liu X, Han W, Sudhof TC (2006) Synapsins regulate use-dependent synaptic plasticity in the calyx of Held by a Ca2+/calmodulin-dependent pathway. Proc Natl Acad Sci U S A 103:2880–2885
Sze CI, Bi H, Kleinschmidt-DeMasters BK, Filley CM, Martin LJ (2000) Selective regional loss of exocytotic presynaptic vesicle proteins in Alzheimer's disease brains. J Neurol Sci 175:81–90
Takahashi K, Saito K, Masuda J, Fujigaki S, Takemura M, Ito H, Seishima M (2007) Changes in neuronal protein expression in LP-BM5-infected mice. Neurosci Lett 422:114–118
Umahara T, Uchihara T, Tsuchiya K, Nakamura A, Iwamoto T, Ikeda K, Takasaki M (2004) 14-3-3 proteins and zeta isoform containing neurofibrillary tangles in patients with Alzheimer's disease. Acta Neuropathol 108:279–286
Wakabayashi H, Yano M, Tachikawa N, Oka S, Maeda M, Kido H (2001) Increased concentrations of 14-3-3 epsilon, gamma and zeta isoforms in cerebrospinal fluid of AIDS patients with neuronal destruction. Clin Chim Acta 312:97–105
Wiley CA, Achim C (1994) Human immunodeficiency virus encephalitis is the pathological correlate of dementia in acquired immunodeficiency syndrome. Ann Neurol 36:673–676
Wood JD, MacMillan JC, Harper PS, Lowenstein PR, Jones AL (1996) Partial characterisation of murine huntingtin and apparent variations in the subcellular localisation of huntingtin in human, mouse and rat brain. Hum Mol Genet 5:481–487
Yi JJ, Ehlers MD (2005) Ubiquitin and protein turnover in synapse function. Neuron 47:629–632
Zhou Y, Schopperle WM, Murrey H, Jaramillo A, Dagan D, Griffith LC, Levitan IB (1999) A dynamically regulated 14-3-3, Slob, and Slowpoke potassium channel complex in Drosophila presynaptic nerve terminals. Neuron 22:809–818
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Source(s) of support: U01MH083507, R01 MH79886, and R01 MH69200.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Gelman, B.B., Nguyen, T.P. Synaptic Proteins Linked to HIV-1 Infection and Immunoproteasome Induction: Proteomic Analysis of Human Synaptosomes. J Neuroimmune Pharmacol 5, 92–102 (2010). https://doi.org/10.1007/s11481-009-9168-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11481-009-9168-0