Abstract
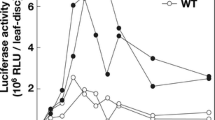
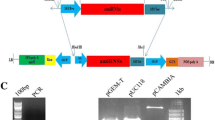
RNA silencing has been shown to function in the plant antivirus defense response, leading to viral RNA degradation induced by vsiRNA-containing RISC cleavage activity. Cucumber mosaic virus (CMV) 3’UTR sequences share a high conservation of nucleotide sequence and secondary structures that are important for CMV replication. Here, in an attempt to simultaneously target the multiple genomic and subgenomic RNAs of CMV for degradation, CMV 3’UTR were used to design hairpin RNA (hpRNA) to transform tobacco (Xanthi. nc) so as to constitutively produce viral siRNAs. Most of the transgenic plants expressing CMV Q strain (Q-CMV, subgroup II strain) RNA3 3’UTR-derived hpRNA showed delayed resistance to Q-CMV infection and exhibited recovery phenotypes. Compared with Q-CMV-inoculated leaves, the upper leaves showed weak or no disease symptoms and a reduced accumulation level of viral RNAs. Together with transient assays, our results indicate that the 3’UTR-derived siRNAs were biologically active in targeting viral RNA for degradation. Recovery resistance in transgenic plants was also observed against subgroup IB strain SD-CMV infection, indicating a broad-spectrum anti-CMV effect of the 3’UTR-based antiviral silencing. Northern blot assays indicated that there was no strong correlation between the degree of resistance and the accumulation level of 3’UTR-derived siRNAs, suggesting that to target a highly structured RNA, such as the CMV 3’UTR, the quantity of siRNAs may not be the only determinant of silencing efficiency. Target RNA secondary structures may also affect target accessibility, siRNA-containing RISC-target recognition and the consequent antiviral effect.
Similar content being viewed by others
References
Goldbach R, Bucher E, Prins M. Resistance mechanisms to plant viruses: An overview. Virus Res, 2003, 92: 207–212
Boonrod K, Galetzka D, Nagy P D, et al. Single-chain antibodies against a plant viral RNA-dependent RNA polymerase confer virus resistance. Nat Biotechnol, 2004, 22: 856–862
Watanabe Y, Ogawa T, Takahashi H, et al. Resistance against multiple plant viruses in plants mediated by a double stranded-RNA specific ribonuclease. FEBS Lett, 1995, 372: 165–168
Masuta C, Tanaka H, Uehara K, et al. Broad resistance to plant viruses in transgenic plants conferred by antisense inhibition of a host gene essential in S-adenosylmethionine-dependent transmethylation reactions. Proc Natl Acad Sci USA, 1995, 92: 6117–6121
Lindbo J A, Silva-Rosales L, Proebsting W M, et al. Induction of a Highly Specific Antiviral State in Transgenic Plants: Implications for Regulation of Gene Expression and Virus Resistance. Plant Cell, 1993, 5: 1749–1759
Palukaitis P, Garcia-Arenal F. Cucumoviruses. Adv Virus Res, 2003, 62: 241–323
Baulcombe D. RNA silencing. Diced defence. Nature, 2001, 409: 295–296
Baulcombe D. RNA silencing in plants. Nature, 2004, 431: 356–363
Schramke V, Sheedy D M, Denli A M, et al. RNA-interference-directed chromatin modification coupled to RNA polymerase II transcription. Nature, 2005, 435: 1275–1279
Baulcombe D. RNA silencing. Trends Biochem Sci, 2005, 30: 290–293
Voinnet O. Induction and suppression of RNA silencing: Insights from viral infections. Nat Rev, 2005, 6: 206–220
Scholthof H B. The Tombusvirus-encoded P19: from irrelevance to elegance. Nat Rev Microbiol, 2006, 4: 405–411
Chellappan P, Vanitharani R, Pita J, et al. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J Virol, 2004, 78: 7465–7477
Molnar A, Csorba T, Lakatos L, et al. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol, 2005, 79: 7812–7818
Pantaleo V, Szittya G, Burgyan J. Molecular bases of viral RNA targeting by viral siRNA programmed RISC. J Virol, 2007, 81: 3797–3806
Guo H S, Garcia J A. Delayed Resistance to plum pox potyvirus mediated by a mutated RNA replicase GENE: Involvement of a gene-silencing mechanism. Mol Plant Microbe Interact, 1997, 10: 160–170
Covey S N, Al-Kaff N S, Langara A, et al. Plants combat infection by gene silencing. Nature, 1997, 385: 781–782
Ratcliff F, Harrison B D, Baulcombe D C. A similarity between viral defense and gene silencing in plants. Science, 1997, 276: 1558–1560
Niu Q W, Lin S S, Reyes J L, et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol, 2006, 24: 1420–1428
Qu J, Ye J, Fang R X. Artificial microRNA-mediated virus resistance in plants. J Virol, 2007, 81: 6690–6699
Simon-Mateo C, Garcia J A. MicroRNA-guided processing impairs plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J Virol, 2006, 80: 2429–2436
Palukaitis P, Roossinck M J, Dietzgen R G, et al. Cucumber mosaic virus. Adv Virus Res, 1992, 41: 281–348
Sugiyama M, Sato H, Karasawa A, et al. Characterization of symptom determinants in two mutants of cucumber mosaic virus Y strain, causing distinct mild green mosaic symptoms in tobacco. Physiol Mol Plant Pathol, 2000, 56: 85–90
Roossinck M J, Zhang L, Hellwald K H. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J Virol, 1999, 73: 6752–6758
Hayes R J, Buck K W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell, 1990, 63: 363–368
Schwinghamer M W, Symons R H. Translation of the four major RNA species of cucumber mosaic virus in plant and animal cell-free systems and in toad oocytes. Virology, 1977, 79: 88–108
Brigneti G, Voinnet O, Li W X, et al. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J, 1998, 17: 6739–6746
Zhang X, Yuan Y R, Pei Y, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev, 2006, 20: 3255–3268
Guo H S, Ding S W. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J, 2002, 21: 398–407
Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet, 2005, 37: 1356–1360
Kwon C S, Chung W I. Differential roles of the 5′ untranslated regions of cucumber mosaic virus RNAs 1, 2, 3 and 4 in translational competition. Virus Res, 2000, 66: 175–185
Rietveld K, Pleij C W, Bosch L. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J, 1983, 2: 1079–1085
Fernandez-Cuartero B, Burgyan J, Aranda M A, et al. Increase in the relative fitness of a plant virus RNA associated with its recombinant nature. Virology, 1994, 203: 373–377
Guo H S, Fei J F, Xie Q, et al. A chemical-regulated inducible RNAi system in plants. Plant J, 2003, 34: 383–392
Ding S W, Rathjen J P, Li W X, et al. Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J Gen Virol, 1995, 76: 459–464
Schob H, Kunz C, Meins F Jr. Silencing of transgenes introduced into leaves by agroinfiltration: A simple, rapid method for investigating sequence requirements for gene silencing. Mol Gen Genet, 1997, 256: 581–585
Llave C, Kasschau K D, Rector M A, et al. Endogenous and silencingssociated small RNAs in plants. Plant Cell, 2002, 14, 1605–1619
Smith N A, Singh S P, Wang M B, et al. Total silencing by intronpliced hairpin RNAs. Nature, 2000, 407: 319–320
Chen Y, Lohuis D, Goldbach R, et al. High frequency induction of RNA-mediated resistance against Cucumber mosaic virus using inverted repeat constructs. Mol Breed, 2004, 14: 215–226
Di Nicola-Negri E, Brunetti A, Tavazza M et al. Hairpin RNA-mediated silencing of plum pox virus P1 and HC-Pro genes for efficient and predictable resistance to the virus. Transgen Res, 2005, 14: 989–994
Kalantidis K, Psaradakis S, Tabler M, et al. The Occurrence of CMV-specific short RNAs in transgenic tobacco expressing virus-derived double-stranded RNA is indicative of resistance to the virus. Mol Plant Microbe Interact, 2002, 15: 826–833
Soards A J, Murphy A M, Palukaitis P, et al. Virulence and differential local and systemic spread of cucumber mosaic virus in tobacco are affected by the CMV 2b protein. Mol Plant Microbe Interact, 2002, 15: 647–653
Wang Y, Tzfira T, Gaba V, et al. Functional analysis of the Cucumber mosaic virus 2b protein: Pathogenicity and nuclear localization. J Gen Virol, 2004, 85: 3135–3147
Ding S W, Li W X, Symons R H. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J, 1995, 14: 5762–5772
Lucy A P, Guo H S, Li W X, et al. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J, 2000, 19: 1672–1680
Du Q S, Duan C G, Zhang Z H, et al. DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J Virol, 2007, 81: 9142–9151
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the State Basic Research Development Program of China (Grant No. 2006CB101906) and National Natural Science Foundation of China (Grant No. 30530500)
About this article
Cite this article
Duan, C., Wang, C. & Guo, H. Delayed resistance to Cucumber mosaic virus mediated by 3’UTR-derived hairpin RNA. Chin. Sci. Bull. 53, 3301–3310 (2008). https://doi.org/10.1007/s11434-008-0440-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-008-0440-0