Abstract
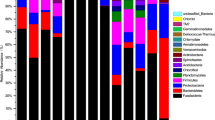
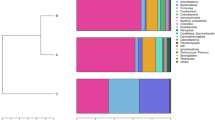
Gut microbiota of four economically important Asian carp species (silver carp, Hypophthalmichthys molitrix; bighead carp, Hypophthalmichthys nobilis; grass carp, Ctenopharyngodon idella; common carp, Cyprinus carpio) were compared using 16S rRNA gene pyrosequencing. Analysis of more than 590,000 quality-filtered sequences obtained from the foregut, midgut and hindgut of these four carp species revealed high microbial diversity among the samples. The foregut samples of grass carp exhibited more than 1,600 operational taxonomy units (OTUs) and the highest alpha-diversity index, followed by the silver carp foregut and midgut. Proteobacteria, Firmicutes, Bacteroidetes and Fusobacteria were the predominant phyla regardless of fish species or gut type. Pairwise (weighted) UniFrac distance-based permutational multivariate analysis of variance with fish species as a factor produced significant association (P<0.01). The gut microbiotas of all four carp species harbored saccharolytic or proteolytic microbes, likely in response to the differences in their feeding habits. In addition, extensive variations were also observed even within the same fish species. Our results indicate that the gut microbiotas of Asian carp depend on the exact species, even when the different species were cohabiting in the same environment. This study provides some new insights into developing commercial fish feeds and improving existing aquaculture strategies.
Similar content being viewed by others
References
Aguilera, E., Yany, G., and Romero, J. (2013). Cultivable intestinal microbiota of yellowtail juveniles (Seriola lalandi) in an aquaculture system. Lat Am J Aquat Res 41, 395–403.
Al-Hisnawi, A., Ringø, E., Davies, S.J., Waines, P., Bradley, G., and Merrifield, D.L. (2015). First report on the autochthonous gut microbiota of brown trout (Salmo trutta Linnaeus). Aquac Res 46, 2962–2971.
Anderson, M.J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26, 32–46.
Austin, B. (2011). Taxonomy of bacterial fish pathogens. Vet Res 42, 20.
Benson, A.K., Kelly, S.A., Legge, R., Ma, F., Low, S.J., Kim, J., Zhang, M., Oh, P.L., Nehrenberg, D., Hua, K., et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107, 18933–18938.
Billard, R., and Berni, P. (2004). Trends in cyprinid polyculture. Cybium 28, 255–261.
Bunn, S.E., Davies, P.M., and Winning, M. (2003). Sources of organic carbon supporting the food web of an arid zone floodplain river. Freshwater Biol 48, 619–635.
Caporaso, J.G., Bittinger, K., Bushman, F.D., DeSantis, T.Z., Andersen, G. L., and Knight, R. (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267.
Caporaso, J.G., Lauber, C.L., Costello, E.K., Berg-Lyons, D., Gonzalez, A., Stombaugh, J., Knights, D., Gajer, P., Ravel, J., Fierer, N., et al. (2011). Moving pictures of the human microbiome. Genome Biol 12, R50.
Claesson, M.J., O’Sullivan, O., Wang, Q., Nikkilä, J., Marchesi, J.R., Smidt, H., de Vos, W.M., Ross, R.P., and O’Toole, P.W. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4, e6669.
Clements, K.D., Angert, E.R., Montgomery, W.L., and Choat, J.H. (2014). Intestinal microbiota in fishes: what’s known and what’s not. Mol Ecol 23, 1891–1898.
Currin, C.A., Levin, L.A., Talley, T.S., Michener, R., and Talley, D. (2011). The role of cyanobacteria in Southern California salt marsh food webs. Mar Ecol 32, 346–363.
David, L.A., Maurice, C.F., Carmody, R.N., Gootenberg, D.B., Button, J.E., Wolfe, B.E., Ling, A.V., Devlin, A.S., Varma, Y., Fischbach, M.A., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563.
Fishery Bureau of the Ministry of Agriculture. (2014). China Fishery Statistical Yearbook. (Beijing: China Agriculture Press).
Gatesoupe, F.J., Huelvan, C., Le Bayon, N., Sévère, A., Aasen, I.M., Degnes, K.F., Mazurais, D., Panserat, S., Zambonino-Infante, J.L., and Kaushik, S.J. (2014). The effects of dietary carbohydrate sources and forms on metabolic response and intestinal microbiota in sea bass juveniles, Dicentrarchus labrax. Aquaculture 422–423, 47–53.
Ghosh, K., Roy, M., Kar, N., and Ringo, E. (2010). Gastrointestinal bacteria in rohu, Labeo Rohita (Actinopterygii: Cypriniformes: Cyprinidae): scanning electron microscopy and bacteriological study. Acta Icth Piscat 40, 129–135.
Gramignoli, R., Green, M.L., Tahan, V., Dorko, K., Skvorak, K.J., Marongiu, F., Zao, W., Venkataramanan, R., Ellis, E.C.S., Geller, D., et al. (2012). Development and application of purified tissue dissociation enzyme mixtures for human hepatocyte isolation. Cell Transplant 21, 1245–1260.
Hamady, M., Lozupone, C., and Knight, R. (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4, 17–27.
Hammer, Ø., Harper, D.A.T., and Ryan, P.D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol Electronica 4, 9.
He, S., Wang, Q., Li, S., Ran, C., Guo, X., Zhang, Z., and Zhou, Z. (2017). Antibiotic growth promoter olaquindox increases pathogen susceptibility in fish by inducing gut microbiota dysbiosis. Sci China Life Sci 60, 1260–1270.
Hooper, L.V., Wong, M.H., Thelin, A., Hansson, L., Falk, P.G., and Gordon, J.I. (2001). Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884.
Jeney, Z., and Jian, Z. (2009). Use and exchange of aquatic resources relevant for food and aquaculture: common carp (Cyprinuscarpio L.). Rev Aquaculture 1, 163–173.
Jiang, Y., Xie, C., Yang, G., Gong, X., Chen, X., Xu, L., and Bao, B. (2011). Cellulase-producing bacteria of Aeromonas are dominant and indigenous in the gut of Ctenopharyngodon idellus (Valenciennes). Aquaculture Res 42, 499–505.
Li, J., Ni, J., Li, J., Wang, C., Li, X., Wu, S., Zhang, T., Yu, Y., and Yan, Q. (2014). Comparative study on gastrointestinal microbiota of eight fish species with different feeding habits. J Appl Microbiol 117, 1750–1760.
Li, T., Long, M., Gatesoupe, F.J., Zhang, Q., Li, A., and Gong, X. (2015). Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb Ecol 69, 25–36.
Li, X.M., Zhu, Y.J., Yan, Q.Y., Ringo, E., and Yang, D.G. (2014). Do the intestinal microbiotas differ between paddlefish (Polyodon spathala) and bighead carp (Aristichthys nobilis) reared in the same pond? J Appl Microbiol 117, 1245–1252.
Li, Z., Xu, L., Liu, W., Liu, Y., Ringø, E., Du, Z., and Zhou, Z. (2015). Protein replacement in practical diets altered gut allochthonous bacteria of cultured cyprinid species with different food habits. Aquacult Int 23, 913–928.
Liu, H., Guo, X., Gooneratne, R., Lai, R., Zeng, C., Zhan, F., and Wang, W. (2016). The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci Rep 6, 24340.
Mandal, S., and Ghosh, K. (2013). Isolation of tannase-producing microbiota from the gastrointestinal tracts of some freshwater fish. J Appl Ichthyol 29, 145–153.
Marungruang, N., Fåk, F., and Tareke, E. (2016). Heat-treated high-fat diet modifies gut microbiota and metabolic markers in apoe -/- mice. Nutr Metab (Lond) 13, 22.
McMurdie, P.J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217.
Mohammed, H.H., and Arias, C.R. (2015). Potassium permanganate elicits a shift of the external fish microbiome and increases host susceptibility to columnaris disease. Vet Res 46, 82.
Nayak, S.K. (2010). Role of gastrointestinal microbiota in fish. Aquacult Res 41, 1553–1573.
Ni, J., Yu, Y., Zhang, T., and Gao, L. (2012). Comparison of intestinal bacterial communities in grass carp, Ctenopharyngodon idellus, from two different habitats. Chin J Ocean Limnol 30, 757–765.
Ni, J., Yan, Q., Yu, Y., and Zhang, T. (2014). Factors influencing the grass carp gut microbiome and its effect on metabolism. FEMS Microbiol Ecol 87, 704–714.
Oksanen, J., Blanchet, F.G., Kindt, R., Legendre, P., Minchin, P.R., O’Hara, R.B., Simpson, G.L., Solymos, P., Stevens, M.H.H., and Wagner, H.H. (2016). vegan: Community Ecology Package. Software.
Park, S.H., Lee, S.I., and Ricke, S.C. (2016). Microbial populations in naked neck chicken ceca raised on pasture flock fed with commercial yeast cell wall prebiotics via an Illumina MiSeq platform. PLoS ONE 11, e0151944.
Pearce, D.A., Gast, C.J., Lawley, B., and Ellis-Evans, J.C. (2003). Bacterioplankton community diversity in a maritime Antarctic lake, determined by culture-dependent and culture-independent techniques. FEMS MicroBiol Ecol 45, 59–70.
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K.S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., Yamada, T., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65.
R Development Core Team (2008) R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Ranjan, R., Rani, A., Metwally, A., McGee, H.S., and Perkins, D.L. (2016). Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun 469, 967–977.
Ray, A.K., Ghosh, K., and Ringø, E. (2012). Enzyme-producing bacteria isolated from fish gut: a review. Aquacult Nutr 18, 465–492.
Ritchie, M.E., Phipson, B., Wu, D., Hu, Y., Law, C.W., Shi, W., and Smyth, G.K. (2015). limma powers differential expression analyses for RNAsequencing and microarray studies. Nucleic Acids Res 43, e47.
Roeselers, G., Mittge, E.K., Stephens, W.Z., Parichy, D.M., Cavanaugh, C. M., Guillemin, K., and Rawls, J.F. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J 5, 1595–1608.
Schwiertz, A., Hold, G.L., Duncan, S.H., Gruhl, B., Collins, M.D., Lawson, P.A., Flint, H.J., and Blaut, M. (2002). Anaerostipes caccae gen. nov., sp nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst Appl Microbiol 25, 46–51.
Shanks, O.C., Kelty, C.A., Archibeque, S., Jenkins, M., Newton, R.J., McLellan, S.L., Huse, S.M., and Sogin, M.L. (2011). Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77, 2992–3001.
Stephens, W.Z., Burns, A.R., Stagaman, K., Wong, S., Rawls, J.F., Guillemin, K., and Bohannan, B.J.M. (2016). The composition of the zebrafish intestinal microbial community varies across development. ISME J 10, 644–654.
Sun, Y., Zhou, L.P., Fang, L.D., Su, Y., and Zhu, W.Y. (2015). Responses in colonic microbial community and gene expression of pigs to a longterm high resistant starch diet. Front Microbiol 6, 877.
Torok, V.A., Ophel-Keller, K., Loo, M., and Hughes, R.J. (2008). Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl Environ Microbiol 74, 783–791.
Tsuchiya, C., Sakata, T., and Sugita, H. (2008). Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett Appl Microbiol 46, 43–48.
Wang, Q., Li, Z., Lian, Y., Du, X., Zhang, S., Yuan, J., Liu, J., and De Silva, S.S. (2016). Farming system transformation yields significant reduction in nutrient loading: case study of Hongze Lake, Yangtze River Basin, China. Aquaculture 457, 109–117.
Warton, D.I., Wright, S.T., and Wang, Y. (2012). Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol Evol 3, 89–101.
Wong, S., and Rawls, J.F. (2012). Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol Ecol 21, 3100–3102.
Wu, L., Wen, C., Qin, Y., Yin, H., Tu, Q., Van Nostrand, J.D., Yuan, T., Yuan, M., Deng, Y., and Zhou, J. (2015). Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol 15, 125.
Wu, S., Gao, T., Zheng, Y., Wang, W., Cheng, Y., and Wang, G. (2010). Microbial diversity of intestinal contents and mucus in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 303, 1–7.
Yan, Q., van der Gast, C.J., and Yu, Y. (2012). Bacterial community assembly and turnover within the intestines of developing zebrafish. PLoS ONE 7, e30603.
Yan, Q., Bi, Y., Deng, Y., He, Z., Wu, L., Van Nostrand, J.D., Shi, Z., Li, J., Wang, X., Hu, Z., et al. (2015). Impacts of the Three Gorges Dam on microbial structure and potential function. Sci Rep 5, 8605.
Yan, Q., Li, J., Yu, Y., Wang, J., He, Z., Van Nostrand, J.D., Kempher, M. L., Wu, L., Wang, Y., Liao, L., et al. (2016). Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ Microbiol 18, 4739–4754.
Ye, L., Amberg, J., Chapman, D., Gaikowski, M., and Liu, W.T. (2014). Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J 8, 541–551.
Zhang, C., Zhang, M., Wang, S., Han, R., Cao, Y., Hua, W., Mao, Y., Zhang, X., Pang, X., Wei, C., et al. (2010). Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 4, 232–241.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31400109, 31372202) and the Youth Innovation Promotion Association, Chinese Academy of Sciences (Y22Z07).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Li, X., Yu, Y., Li, C. et al. Comparative study on the gut microbiotas of four economically important Asian carp species. Sci. China Life Sci. 61, 696–705 (2018). https://doi.org/10.1007/s11427-016-9296-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-016-9296-5