Abstract
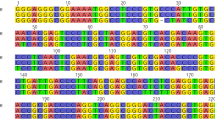
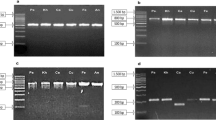
Based on variable nuclear and/or organellar DNA sequences among vastly divergent species as well as morphologically indistinguishable species, DNA barcoding is widely applicable in species identification, biodiversity studies, forensic analyses, and authentication of medicinal plants. The roots of Astragalus membranaceus and A. membranaceus var. mongholica are commonly used as Radix Astragali in several Asian countries, including China, Japan, and Korea. However, in addition to the two species recorded in the Chinese Pharmacopoeia, there are twenty-three species from different genera including Astragalus, Oxytropis, Hedysarum, and Glycyrrhiza, which have been used as adulterants not only in trading markets but also by the herbal medicine industry. Therefore, a simple, reliable, and accurate classification method is important for distinguishing authentic Radix Astragali from its adulterants. In this study, we acquired data for 37 samples from four related genera within the family Fabaceae. Then we compared four candidate DNA barcoding markers using ITS, matK, rbcL, and coxI sequences from nuclear, chloroplast, and mitochondrial genomes, all commonly used for plants to identify genetic variations among genera, intraspecies, and interspecies. We observed higher divergences among genera and interspecies for ITS, which have the average Kimura 2-parameter distances of 4.5% and 14.1%, respectively, whereas matK was found to have sufficient divergence at the intraspecific level. Moreover, two indels detected in the matK sequence are useful for PCR studies in distinguishing Radix Astragali from its adulterants. This study suggests that the combined barcoding regions of ITS and matK are superior barcodes for Radix Astragali and further studies should focus on evaluating the applicability and accuracy of such combined markers for a wide range of traditional Chinese herbs.
Similar content being viewed by others
References
Pharmacopoeia Of The People’s Republic Of China. Beijing: Chemical Industry Press. 2010
Cui X M, Lo C K, Yip K L, et al. Authentication of Panax notoginseng by 5S-rRNA spacer domain and random amplified polymorphic DNA (RAPD) analysis. Planta Med, 2003, 69: 584–586, 1:CAS:528:DC%2BD3sXlsFGisbo%3D, 10.1055/s-2003-40632, 12865989
Sarwat M, Das S, Srivastava P S. Analysis of genetic diversity through AFLP, SAMPL, ISSR and RAPD markers in Tribulus terrestris, a medicinal herb. Plant Cell Rep, 2008, 27: 519–528, 1:CAS:528:DC%2BD1cXhvFygt7Y%3D, 10.1007/s00299-007-0478-5, 18074139
Dangi R S, Lagu M D, Choudhary L B, et al. Assessment of genetic diversity in Trigonella foenum-graecum and Trigonella caerulea using ISSR and RAPD markers. BMC Plant Biol, 2004, 4: 13, 10.1186/1471-2229-4-13, 15285785
Yip P Y, Kwan H S. Molecular identification of Astragalus membranaceus at the species and locality levels. J Ethnopharmacol, 2006, 106: 222–229, 1:CAS:528:DC%2BD28XltFOrtb4%3D, 10.1016/j.jep.2005.12.033, 16442761
Zhang Y B, Ngan F N, Wang Z T, et al. Random primed polymerase chain reaction differentiates Codonopsis pilosula from different localities. Planta Med, 1999, 65: 157–160, 10.1055/s-1999-14058, 17260247
Ni X, Huang Y, Wu L, et al. Genetic diversity of the endangered Chinese endemic herb Primulina tabacum (Gesneriaceae) revealed by amplified fragment length polymorphism (AFLP). Genetica, 2006, 127: 177–183, 1:CAS:528:DC%2BD28XntVCqtLo%3D, 10.1007/s10709-005-3227-0, 16850222
Wang C Z, Li P, Ding J Y, et al. Simultaneous identification of Bulbus Fritillariae cirrhosae using PCR-RFLP analysis. Phytomedicine, 2007, 14: 628–632, 1:CAS:528:DC%2BD2sXhtFemsr3O, 10.1016/j.phymed.2006.09.008, 17336047
Cannon C H, Kua C S, Zhang D, et al. Assembly free comparative genomics of short-read sequence data discovers the needles in the haystack. Mol Ecol, 2010, 19: S147–S161, 10.1111/j.1365-294X.2009.04484.x
Hebert P D, Ratnasingham S, deWaard J R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci, 2003, 270: S96–S99, 1:CAS:528:DC%2BD3sXns1Smsbo%3D, 10.1098/rsbl.2003.0025, 12952648
Saunders G W. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philos Trans R Soc Lond B Biol Sci, 2005, 360: 1879–1888, 1:CAS:528:DC%2BD2MXhtlSjsrnO, 10.1098/rstb.2005.1719, 16214745
Nassonova E, Smirnov A, Fahrni J, et al. Barcoding amoebae: comparison of SSU, ITS and COI genes as tools for molecular identification of naked lobose amoebae. Protist, 2010, 161: 102–115, 1:CAS:528:DC%2BC3cXktlOnsrY%3D, 10.1016/j.protis.2009.07.003, 19819756
Zhang J, Wang J, Xia T, et al. DNA barcoding: species delimitation in tree peonies. Sci China Ser C-Life Sci, 2009, 52: 568–578, 1:CAS:528:DC%2BD1MXnvVWlsLg%3D, 10.1007/s11427-009-0069-5
Zhu Y J, Chen S L, Yao H, et al. DNA barcoding the medicinal plants of the genus Paris. Acta Pharm Sin, 2010, 45: 376–382, 1:CAS:528:DC%2BC3cXhtVKls7nK
Shi L C, Liang Z S, Han J P, et al. Screening potential DNA barcode regions in Rhododendron. World Sci Tech/Modern Trad Chin Med, 2009, 11: 54–57
Chen S, Yao H, Han J, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One, 2010, 5: e8613, 10.1371/journal.pone.0008613, 20062805
Yao H, Song J Y, Ma X Y, et al. Identification of Dendrobium species by a candidate DNA barcode sequence: the chloroplast psbA-trnH intergenic region. Planta Med, 2009, 75: 667–669, 1:CAS:528:DC%2BD1MXmsFWhu7g%3D, 10.1055/s-0029-1185385, 19235685
Song J, Yao H, Li Y, et al. Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique. J Ethnopharmacol, 2009, 124: 434–439, 1:CAS:528:DC%2BD1MXptVSisbg%3D, 10.1016/j.jep.2009.05.042, 19505556
Selvaraj D, Sarma R K, Sathishkumar R. Phylogenetic analysis of chloroplast matK gene from Zingiberaceae for plant DNA barcoding. Bioinformation, 2008, 3: 24–27, 19052662
Zhou R F, Liu P X, Tan M. Effect of Astragalus mongholicus injection on proliferation and apoptosis of hormone sensitive (MCF-7) breast cancer cell lines with physiological dose E2. Zhong Yao Cai, 2009, 32: 744–747, 1:CAS:528:DC%2BD1MXhsFWgurjE, 19771851
Hu Y W, Liu C Y, Du CM, et al. Induction of apoptosis in human hepatocarcinoma SMMC-7721 cells in vitro by flavonoids from Astragalus complanatus. J Ethnopharmacol, 2009, 123: 293–301, 1:CAS:528:DC%2BD1MXlsVKhsrw%3D, 10.1016/j.jep.2009.03.016, 19429375
Cassileth B R, Rizvi N, Deng G, et al. Safety and pharmacokinetic trial of docetaxel plus an Astragalus-based herbal formula for non-small cell lung cancer patients. Cancer Chemother Pharmacol, 2009, 65: 67–71, 1:CAS:528:DC%2BD1MXhtFegu7zJ, 10.1007/s00280-009-1003-z, 19421753
Clement-Kruzel S, Hwang S A, Kruzel M C, et al. Immune modulation of macrophage pro-inflammatory response by goldenseal and Astragalus extracts. J Med Food, 2008, 11: 493–498, 1:CAS:528:DC%2BD1cXhtFCitLbN, 10.1089/jmf.2008.0044, 18800897
Chen J, Gui D, Chen Y, et al. Astragaloside IV improves high glucose-induced podocyte adhesion dysfunction via alpha3beta1 integrin upregulation and integrin-linked kinase inhibition. Biochem Pharmacol, 2008, 76: 796–804, 1:CAS:528:DC%2BD1cXhtVyku7fF, 10.1016/j.bcp.2008.06.020, 18652804
Li R J, Qiu S D, Chen H X, et al. The immunotherapeutic effects of Astragalus polysaccharide in type 1 diabetic mice. Biol Pharm Bull, 2007, 30: 470–476, 1:CAS:528:DC%2BD2sXkvVSntL4%3D, 10.1248/bpb.30.470, 17329840
Cho W C, Leung K N. In vitro and in vivo anti-tumor effects of Astragalus membranaceus. Cancer Lett, 2007, 252: 43–54, 1:CAS:528:DC%2BD2sXlsF2ks7k%3D, 10.1016/j.canlet.2006.12.001, 17223259
Shen P, Liu M H, Ng T Y, et al. Differential effects of isoflavones, from Astragalus membranaceus and Pueraria thomsonii, on the activation of PPARalpha, PPARgamma, and adipocyte differentiation in vitro. J Nutr, 2006, 136: 899–905, 1:CAS:528:DC%2BD28XjtVOks7o%3D, 16549448
Zhang J, Xie X, Li C, et al. Systematic review of the renal protective effect of Astragalus membranaceus (root) on diabetic nephropathy in animal models. J Ethnopharmacol, 2009, 126: 189–196, 1:CAS:528:DC%2BD1MXhtlGjt73K, 10.1016/j.jep.2009.08.046, 19735713
Wang S, Li J, Huang H, et al. Anti-hepatitis B virus activities of astragaloside IV isolated from radix Astragali. Biol Pharm Bull, 2009, 32: 132–135, 10.1248/bpb.32.132, 19122295
Wu Z. XINHUABENCAOGANGYAO. Shanghai: Shanghai Technology Publishing Company. 1991. Vol 2: 94–149
Sun S S, Lin Z, Chu C C. Identification and investigation of Radix Astragali. Chinese. Pharm J, 1990, 25: 643–647
Gang J, Guo P J, An investigation on the resources of Astragalus herbal products in Qinghai. J Chinese Med Mater, 1993, 16: 15–19
Zhao M, Duan J A, Huang W Z, et al. Study on the status and resources of Astragalus medicinal plants. Chinese Wild Plant Res, 2000, 19: 5–9
Zhao Y Z. Investigation the source and distribution of Radix Astragli. Chinese Trad Herbal Drugs, 2004, 35: 1189–1190
Zhang Z Z, et al. Characterization and recognition key components in Astragalus membranaceu. Yao Xue Xue Bao, 2001, 36: 523–527, 1:STN:280:DC%2BD3s%2FnsFSguw%3D%3D, 12585085
Ma X Q, Shi Q, Duan J A, et al. Chemical analysis of Radix Astragali (Huangqi) in China: a comparison with its adulterants and seasonal variations. J Agric Food Chem, 2002, 50: 4861–4866, 1:CAS:528:DC%2BD38XltlaisL4%3D, 10.1021/jf0202279, 12166972
Tanaka K, Tamura T, Fukuda S, et al. Quality evaluation of Astragali Radix using a multivariate statistical approach. Phytochemistry, 2008, 69: 2081–2087, 1:CAS:528:DC%2BD1cXnvVGrtbc%3D, 10.1016/j.phytochem.2008.04.016, 18534641
Na H J, Um J Y, Kim S C, et al. Molecular discrimination of medicinal Astragali radix by RAPD analysis. Immunopharmacol Immunotoxicol, 2004, 26: 265–272, 1:CAS:528:DC%2BD2cXjvFKms7k%3D, 10.1081/IPH-120037723, 15209362
Chen G, Wang X L, Wong W S, et al. Application of 3′ untranslated region (UTR) sequence-based amplified polymorphism analysis in the rapid authentication of Radix Astragali. J Agric Food Chem, 2005, 53: 8551–8556, 1:CAS:528:DC%2BD2MXhtVGqsL7O, 10.1021/jf051334g, 16248552
Qian D, Huang L, Cui G, et al. Study on genetic relationship of Astragalus membranaceus var mongholicus in different producing area using SRAP. Zhongguo Zhong Yao Za Zhi, 2009, 34: 382–385, 1:CAS:528:DC%2BD1MXlslSrsrk%3D, 19459294
Ma X Q, Duan J A, Zhu D Y, et al. Species identification of Radix Astragali (Huangqi) by DNA sequence of its 5S-rRNA spacer domain. Phytochemistry, 2000, 54: 363–368, 1:CAS:528:DC%2BD3cXkslOgtLc%3D, 10.1016/S0031-9422(00)00111-4, 10897476
Thompson J D, Gibson T J, Higgins D G. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinform, 2002, Chapter 2: Unit 2 3
Zhao Y Z. Taxonomy and floristic geographical distribution of the Chinese medicinal Huangqi. Bull Botan Res, 2006, 26: 532–538
Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol, 2007, 24: 1596–1599, 1:CAS:528:DC%2BD2sXpsVGrsL8%3D, 10.1093/molbev/msm092, 17488738
Author information
Authors and Affiliations
Corresponding author
Additional information
Contributed equally to this work
Rights and permissions
About this article
Cite this article
Guo, H., Wang, W., Yang, N. et al. DNA barcoding provides distinction between Radix Astragali and its adulterants. Sci. China Life Sci. 53, 992–999 (2010). https://doi.org/10.1007/s11427-010-4044-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-010-4044-y