Abstract
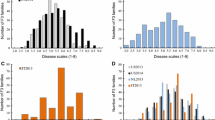
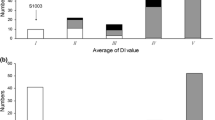
The cucumber lines, S94 (Northern China open-field type, powdery mildew (PM) susceptible) and S06 (European greenhouse type, PM resistant), and their F6:7 populations were used to investigate PM resistance under seedling spray inoculation in 2005/Autumn and 2006/Spring. QTL analysis was undertaken based on a constructed molecular linkage map of the corresponding F6 population using composite interval mapping. A total of four QTLs (pm1.1, pm2.1, pm4.1 and pm6.1) for PM resistance were identified and located on LG 1, 2, 4 and 6, respectively, explaining 5.2%–21.0% of the phenotypic variation. Three consistent QTLs (pm1.1, pm2.1 and pm4.1) were detected under the two test conditions. The QTL pm6.1 was only identified in 2005/Autumn. The total phenotypic variation explained by the QTLs was 52.0% and 42.0% in 2005/Autumn and 2006/Spring, respectively. Anchor markers tightly linked to those loci (<5 cM) could lay a basis for both molecular marker-assisted breeding and map-based gene cloning of the PM-resistance gene in cucumber.
Similar content being viewed by others
References
Lv P K, Li M Y, Wu J W. The disease of cucumber. In: Wei W, ed. China Vegetable Plant Diseases and Insect Pests Primitive Colors Pictures (in Chinese). Beijing: China Agriculture Press, 1996, 66–76
Feng D. The recent progress in resistance powdery mildew breeding of cucurbitaceous. China Veget (in Chinese), 1996, 1: 55–59
Morishita M, Sugiyama K, Saito T, et al. An improved evaluation method for screening and selecting powdery mildew resistant cultivars and lines of cucumber (Cucumis sativus L.). J Jpn Soc Hortic Sci, 2002, 71: 94–100, 10.2503/jjshs.71.94
Hou F, Lv S Z, Ma D H. Powdery mildew of cucumber. In: Yin D, ed. Cucumber (in Chinese). Tianjin: Tianjin Technology Press, 1999. 756–778
McGrath M T. Fungicide resistance in cucurbit powdery mildew: experiences and challenges. Plant Disease, 2001, 85(3): 236–245, 10.1094/PDIS.2001.85.3.236
Mao A Y, Zhang F, Zhang H L, et al. Inheritance of resistance to powdery mildew in two cucumber varieties. Chin Agric Sci Bull (in Chinese), 2005, 21(6): 302–304
Munger H M. Dominant genes for resistance to powdery mildew in cucumber. Cucurbit Genet Coop, 1979, 2: 10–15
Morishita M, Sugiyama K, Saitot S, et al. Powdery mildew resistance in cucumber. Jpn Agr Res Q, 2003, 37(1): 7–14
Sakata Y, Kubo N, Morishita M, et al. QTL analysis of powdery mildew resistance in cucumber (Cucumis sativus L.). Theor Appl Genet, 2006, 112(2): 243–250, 16240105, 10.1007/s00122-005-0121-1, 1:CAS:528:DC%2BD2MXhtlajsrzE
Zhang G H, Du S L, Wang M, et al. AFLP markers of cucumber powdery mildew resistance-related gene. Acta Hort Sin (in Chinese), 2004, 31(2): 189–192
Lander E S, Green P, Abrahamson J, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural population. Genomics, 1987, 1: 174–181, 3692487, 10.1016/0888-7543(87)90010-3, 1:CAS:528:DyaL1cXhsVCksrk%3D
Yuan X J, Li X Z, Pan J S, et al. Genetic linkage map construction and location of QTLs for fruit-related traits in cucumber. Plant Breeding, 2008, 127(2): 180–188, 10.1111/j.1439-0523.2007.01426.x, 1:CAS:528:DC%2BD1cXltl2nu7g%3D
Kong Q, Xiang C, Yu Z. Development of EST-SSRs in Cucumis sativus from sequence database. Mol Ecol Notes, 2006, 6(4): 1234–1236, 10.1111/j.1471-8286.2006.01500.x, 1:CAS:528:DC%2BD2sXmvVSntg%3D%3D
Robbins M D. Molecular marker development, QTL pyramiding, and comparative analysis of phenotypic and marker-assisted selection in cucumber. Doctor Dissertation. University of Wisconsin-Madison, 2006
Yuan X J, Pan J S, Cai R, et al. Genetic mapping and QTL analysis of fruit and flower related traits in cucumber (Cucumis sativus L.) using recombinant inbred lines. Euphytica, 2008, online DOI 10.1007/s10681-008-9722-5
Block C C, Reitsma K R. Powdery mildew resistance in the U.S. National plant germplasm system cucumber collection. HortScience, 2005, 40(2): 416–420
Levine D M, Ramsey P P, Smidt R K. Applied Statistics for Engineers and Scientists: Using Microsoft Excel and MINITABLE. Upper Saddle River, NJ: Prentice-Hall Inc., 2001
Zeng Z B. Theoretical basis of separation of multiple linked gene effects on mapping quantitative trait loci. Proc Natl Acad Sci USA, 1993, 90: 10972–10976, 8248199, 10.1073/pnas.90.23.10972, 1:CAS:528:DyaK2cXkslWlsQ%3D%3D
Epinat C, Pitrat M, Bertrand F. Genetic analysis of resistance of five melon lines to powdery mildews. Euphytica, 1993, 65(2):135–144, 10.1007/BF00022575
Cohen R. A leaf disk assay for detection of resistance of melons to Sphaerotheca fuligiea race 1. Plant Disease, 1993, 77(5): 513–517
Chen H, Wang S, Xing Y, et al. Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc Natl Acad Sci USA, 2003, 100(5): 2544–2549, 12601171, 10.1073/pnas.0437898100, 1:CAS:528:DC%2BD3sXitVaisL4%3D
Cherif M, Harrabi M. Transgressive segregation for resistance to Pyrenophora teres in barley. Plant Pathology, 1993, 42(4): 617–21, 10.1111/j.1365-3059.1993.tb01542.x
Thomas W T B, Powell W, Waugh R, et al. Detection of quantitative trait loci for agronomic, yield, grain and disease characters in spring barley (Hordeum vulgare L.). Theor Appl Genet, 1995, 91(6–7): 1037–1047, 1:CAS:528:DyaK28XmtVChuw%3D%3D
Darvishzadeh R, Poormohammad S, Dechamp G, et al. Quantitative trait loci associated with isolate specific and isolate nonspecific partial resistance to Phoma macdonaldii in sunflower. Plant Pathology, 2007, 56(5): 855–861, 10.1111/j.1365-3059.2007.01620.x, 1:CAS:528:DC%2BD2sXhtF2rurrP
Crute I R, Pink D. Genetics and utilization of pathogen resistance in plants. Plant Cell, 1996, 8(10): 1745–1755, 10.1105/tpc.8.10.1747
Miguel A B, Mark J C, Douglas E H, et al. Map position of 47 Arabidopsis sequence with sequence similarity to disease resistance genes. Plant J, 1997, 12(5): 1197–1211, 10.1046/j.1365-313X.1997.12051197.x
Grumet R, Kabelka E, McQueen S, et al. Characterization of sources of resistance to the watermelon strain of Papaya ringspot virus in cucumber: Allelism and co-segregation with other potyvirus resistances. Theor Appl Genet, 2000, 101(3): 463–472, 10.1007/s001220051504
Perchepied L, Bardin M, Dogimont C, et al. Relationship between loci conferring downy mildew and powdery mildew resistance in melon assessed by quantitative trait loci mapping. Phytopathology, 2005, 95(5): 556–565, 18943322, 10.1094/PHYTO-95-0556, 1:CAS:528:DC%2BD2MXktlOjtbY%3D
Young N D. QTL mapping and quantitative disease resistance in plants. Annu Rev Phytopathol, 1996, 34: 479–501, 15012553, 10.1146/annurev.phyto.34.1.479, 1:CAS:528:DyaK28XlsFOhsb8%3D
Fan Z, Robbins M D, Staub J E. Population development by phenotypic selection with subsequent marker-assisted selection for line extraction in cucumber (Cucumis sativus L.). Theor Appl Genet, 2006, 112(5): 843–885, 16397790, 10.1007/s00122-005-0186-x, 1:CAS:528:DC%2BD28XitVanurw%3D
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by grant from the Science and Technology Commission of Shanghai Municipality (Grant No. 06JC14035) and Shanghai Leading Academic Discipline Project (Grant No. B209)
Rights and permissions
About this article
Cite this article
Liu, L., Cai, R., Yuan, X. et al. QTL molecular marker location of powdery mildew resistance in cucumber (Cucumis sativus L.). SCI CHINA SER C 51, 1003–1008 (2008). https://doi.org/10.1007/s11427-008-0110-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-008-0110-0