Abstract
Purpose
Rhizosphere soil bacterial communities are crucial to plant growth, health, and stress resistance. In order to detect how bacterial communities associated with the rhizosphere of phylogenetically related plant species vary in terms of composition, function, and diversity, we investigated the rhizosphere bacterial community structure of two perennial shrub species, Caragana jubata and Caragana roborovskyi, under natural field conditions in northwest China and analyzed the influence of soil properties and environmental factors.
Materials and methods
Eighteen root samples, eight for C. jubata, and ten for C. roborovskyi, along with any adherent soil particles, were collected from multiple sites in northwest China. The rhizosphere soil was washed from the roots, and bacterial communities were analyzed using Illumina MiSeq sequencing of 16S rRNA gene amplicons. Then, α-diversity and β-diversity were calculated using QIIME.
Results and discussion
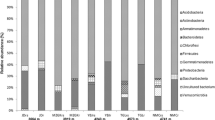
Across species, Proteobacteria (29 %), Actinobacteria (15 %), Chloroflexi (10 %), Acidobacteria (10 %), Bacteroidetes (8 %), Firmicutes (8 %), Planctomycetes (7 %), Gemmatimonadetes (4 %), and Verrucomicrobia (3 %) were the most abundant phyla in the rhizosphere of C. jubata and C. roborovskyi. However, principal co-ordinates analysis indicated strong interspecific patterns of bacterial rhizosphere communities. Further, the richness of Proteobacteria, Acidobacteria, Bacteroidetes, Verrucomicrobia, Firmicutes, and Nitrospirae was significantly higher in the rhizosphere of C. jubata compared with C. roborovskyi, while the opposite was found for Actinobacteria and Cyanobacteria. However, the Shannon index showed no significant difference in α-diversity between C. jubata and C. roborovskyi. Distance-based redundancy analysis indicated that soil properties and environmental factors exerted strong influences on the structure of the rhizosphere bacterial community and explained 47 and 46 % of community variances between samples, respectively.
Conclusions
Our results showed strong interspecific clustering of the bacterial rhizosphere communities of C. roborovskyi and C. jubata. Altitude explained most of the variation in the composition of bacterial rhizosphere communities of C. roborovskyi and C. jubata, followed by soil pH, water content, organic matter content, total nitrogen content, and mean annual rainfall.





Similar content being viewed by others
References
Andrew DR, Fitak RR, Munguia-Vega A, Racolta A, Martinson VG, Dontsova K (2012) Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl Environ Microbiol 78(2):7527–7537
Aira M, Gómez-Brandón M, Lazcano C, Bååth E, Domínguez J (2010) Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol Biochem 42(12):2276–2281
Badri DV, Weir TL, Van Der Lelie D, Vivanco JM (2009) Rhizosphere chemical dialogues: plant–microbe interactions. Curr Opin Biotech 20(6):642–650
Bais HP, Broeckling CD, Vivanco JM (2008) Root exudates modulate plant microbe interactions in the rhizosphere. In: Karlovsky P (ed) Secondary metabolites in soil ecology, soil biology 14. Springer, Berlin, Heidelberg, pp 241–252
Batten KM, Scow KM, Davies KF, Harrison SP (2006) Two invasive plants alter soil microbial community composition in serpentine grasslands. Biol Invasions 8(2):217–230
Berendsen RL, Pieterse CM, Bakker PA (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17(8):478–486
Bhardwaj D, Ansari MW, Sahoo RK, Tuteja N (2014) Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb Cell Fact 13:66–76
Broughton WJ, Zhang F, Perret X, Staehelin C (2003) Signals exchanged between legumes and Rhizobium: agricultural uses and perspectives. Plant Soil 252(1):129–137
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chaparro JM, Badri DV, Vivanco JM (2014) Rhizosphere microbiome assemblage is affected by plant development. The ISME journal 8:790–803
Chen Z, Wu W, Shao X, Li L, Guo Y, Ding G (2015) Shifts in abundance and diversity of soil ammonia-oxidizing bacteria and archaea associated with land restoration in a semi-arid ecosystem. PLoS One 10:e0132879
Chen XR, Li YP, Li QY, Xing YP, Liu BB, Tong YH, Xu JY (2016) SCR96, a small cysteine-rich secretory protein of Phytophthora cactorum, can trigger cell death in the Solanaceae and is important for the pathogenicity and oxidative stress tolerance. Mol Plant Pathol 17(4):577–87
de Groot A, Dulermo R, Ortet P, Blanchard L, Guérin P, Fernandez B, Vacherie B, Dossat C, Jolivet E, Siguier P, Chandler M, Barakat M, Dedieu A, Barbe V, Heulin T, Sommer S, Achouak W, Armengaud J (2009) Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet 5(3):e1000434
de Vrieze J (2015) The littlest farmhands. Science 349(6249):680–683
Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS, Knight R, Gilbert JA, McCulley RL (2013) Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science 342(6158):621–624
Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109(52):21390–21395
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88(6):1354–1364
Fuerst JA (1995) The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiol 141(7):1493–1506
Gao LF, Hu ZA, Wang HX (2002) Genetic diversity of rhizobia isolated from Caragana intermedia in Maowusu sandland, north of China. Lett Appl Microbiol 35(4):347–352
Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils. Environ Microbiol 13(6):1642–1654
Hannula SE, de Boer W, van Veen J (2012) A 3-year study reveals that plant growth stage, season and field site affect soil fungal communities while cultivar and GM-trait have minor effects. PLoS One 7(4):e33819
Hou X, Liu JE, Zhao YZ, Zhao LQ (2006) Interspecific relationships of Caragana microphylla, C. davazamcii and C. korshinskii (Leguminosae) based on ITS and trnL-F data sets. Acta Phytotaxon Sin 44:126–134
Innes L, Hobbs PJ, Bardgett RD (2004) The impacts of individual plant species on rhizosphere microbial communities in soils of different fertility. Biol Fertil Soils 40(1):7–13
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72(3):1719–1728
Ji Z, Yan H, Cui Q, Wang E, Chen W, Chen W (2015) Genetic divergence and gene flow among Mesorhizobium strains nodulating the shrub legume Caragana. Syst Appl Microbiol 38(3):176–183
Khare E, Arora NK (2010) Effect of indole-3-acetic acid (IAA) produced by Pseudomonas aeruginosa in suppression of charcoal rot disease of chickpea. Curr Microbiol 61(1):64–68
Kowalchuk GA, Hol WHG, Van Veen JA (2006) Rhizosphere fungal communities are influenced by Senecio jacobaea pyrrolizidine alkaloid content and composition. Soil Biol Biochem 38(9):2852–2859
Lauber C, Knight R, Hamady M, Fierer N (2009) Soil pH as a predictor of soil bacterial community structure at the continental scale: a pyrosequencing-based assessment. Appl Environ Microbiol 75:5111–5120
Lu YL, Chen WF, Wang ET, Guan SH, Yan XR, Chen WX (2009) Genetic diversity and biogeography of rhizobia associated with Caragana species in three ecological regions of China. Syst Appl Microbiol 32(5):351–361
Ma CC, Gao YB, Guo HY, Wang JL, Wu JB, Xu JS (2008) Physiological adaptations of four dominant Caragana species in the desert region of the Inner Mongolia Plateau. J Arid Environ 72(3):247–254
Madden NM, Southard RJ, Mitchell JP (2009) Soil water content and soil disaggregation by disking affects PM10 emissions. J Environ Qual 38:36–43
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963
Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Øvreås L, Reysenbach AL, Smith VH, Staley JT (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112
McLean EO (1982) Soil pH and lime requirement. In: Page AL (ed) Methods of soil analysis, part 2. American Soc. Agronomy, Madison, pp 199–224
Meng QX, Niu Y, Niu XW, Roubin RH, Hanrahan JR (2009) Ethnobotany, phytochemistry and pharmacology of the genus Caragana used in traditional Chinese medicine. J Ethnopharmacol 124(3):350–368
Micallef SA, Shiaris MP, Colon-Carmona A (2009) Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot 60(6):1729–1742
Mougel C, Offre P, Ranjard L, Corberand T, Gamalero E, Robin C, Lemanceau P (2006) Dynamic of the genetic structure of bacterial and fungal communities at different developmental stages of Medicago truncatula Gaertn. cv. Jemalong line J5. New Phytol 170(1):165–175
Niu XF, Li YM, Hu H, Liu X, Qi L (2013) Chemical constituents from Caragana tangutica. Biochem Syst Ecol 51:288–290
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110(16):6548–6553
Rodriguez RJ, White JF Jr, Arnold AE, Redman RS (2009) Fungal endophytes: diversity and functional roles. New Phytol 182(2):314–330
Shakya M, Quince C, Campbell JH, Yang ZK, Schadt CW, Podar M (2013) Comparative metagenomic and rRNA microbial diversity characterization using archaeal and bacterial synthetic communities. Environ Microbiol 15(6):1882–1899
Shen CC, Xiong JB, Zhang HY, Feng YZ, Lin XG, Li XY, Liang WJ, Chu HY (2013) Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211
Sinha RK, Valani D, Chauhan K, Agarwal S (2010) Embarking on a second green revolution for sustainable agriculture by vermiculture biotechnology using earthworms: reviving the dreams of Sir Charles Darwin. J Agr Biot Sust Dev 2(7):113–128
Siqueira GM, Dafonte JD, Armesto MV, França e Silva EF (2013) Using multivariate geostatistics to assess patterns of spatial dependence of apparent soil electrical conductivity and selected soil properties. Sci World J 2014:712403–712403
Šmilauer P, Lepš J (2014) Multivariate analysis of ecological data using CANOCO 5. Cambridge University Press
Spence CA, Raman V, Donofrio NM, Bais HP (2014) Global gene expression in rice blast pathogen Magnaporthe oryzae treated with a natural rice soil isolate. Planta 239(1):171–85
Tedersoo L, Bahram M, Põlme S et al (2014) Fungal biogeography. Global diversity and geography of soil fungi. Science 346(6213):1256688
Teixeira L, Peixoto RS, Cury JC, Sul WJ, Pellizari VH, Tiedje J, Rosado AS (2010) Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J 4:989–1001
Turner TR, James EK, Poole PS (2013) The plant microbiome. Genome Biol 14(6):209
Uroz S, Buée M, Murat C, Frey-Klett P, Martin F (2010) Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2(2):281–288
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267
Weisskopf L, Abou-Mansour E, Fromin N, Tomasi N, Santelia D, Edelkott I, Neumann G, Aragno M, Tabacchi R, Martinoia E (2006) White lupin has developed a complex strategy to limit microbial degradation of secreted citrate required for phosphate acquisition. Plant Cell Environ 29(5):919–927
Yan XR, Chen WF, Fu JF, Lu YL, Xue CY, Sui XH, Li Y, Wang ET, Chen WX (2007) Mesorhizobium spp. are the main microsymbionts of Caragana spp. grown in Liaoning Province of China. FEMS Microbiol Lett 271:265–273
Yuan YL, Si GC, Wang J, Luo TX, Zhang GX (2014) Bacterial community in alpine grasslands along an altitudinal gradient on the Tibetan Plateau. FEMS Microbiol Ecol 87:121–132
Zhang ML, Landiges YP, Nelson GS (2002) TASS and an analysis of the genus Caragana. Acta Bot Sin 44:1213–1218
Zhang ML, Fritsch PW, Cruz BC (2009) Phylogeny of Caragana (Fabaceae) based on DNA sequence data from rbcL, trnS–trnG, and ITS. Mol Phylogenet Evol 50(3):547–559
Zhao YZ (2009) Classification and its floristic geography of Caragana Fabr. in the word. China. Inner Mongolia University Press, Inner Mongolia, China
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (Nos. 31260166, 31360185, and 31560345).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Chengrong Chen
Xiaofan Na and Ting Ting Xu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1440 kb)
Rights and permissions
About this article
Cite this article
Na, X., Xu, T.T., Li, M. et al. Bacterial diversity in the rhizosphere of two phylogenetically closely related plant species across environmental gradients. J Soils Sediments 17, 122–132 (2017). https://doi.org/10.1007/s11368-016-1486-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-016-1486-2