Abstract
Hospital wastewater contains a variety of human antibiotics and pathogens, which makes the treatment of hospital wastewater essential. However, there is a lack of research on these pollutants at hospital wastewater treatment plants. In this study, the characteristics and removal of antibiotics and antibiotic resistance genes (ARGs) in the independent treatment processes of hospitals of different scales (primary hospital, H1; secondary hospital, H2; and tertiary hospital, H3) were investigated. The occurrence of antibiotics and ARGs in wastewater from three hospitals varied greatly. The first-generation cephalosporin cefradine was detected at a concentration of 2.38 μg/L in untreated wastewater from H1, while the fourth-generation cephalosporin cefepime had the highest concentration, 540.39 μg/L, at H3. Ofloxacin was detected at a frequency of 100% and had removal efficiencies of 44.2%, 51.5%, and 81.6% at H1, H2, and H3, respectively. The highest relative abundances of the β-lactam resistance gene blaGES-1 (1.77×10−3 copies/16S rRNA), the quinolone resistance gene qnrA (8.81×10−6 copies/16S rRNA), and the integron intI1 (1.86×10−4 copies/16S rRNA) were detected in the treated wastewater. The concentrations of several ARGs were increased in the treated wastewater (e.g. blaOXA-1, blaOXA-10, and blaTEM-1). Several pathogenic or opportunistic bacteria (e.g. Acinetobacter, Klebsiella, Aeromonas, and Pseudomonas) were observed at high relative abundances in the treated wastewater. These results suggested the co-occurrence of antibiotics, ARGs, and antibiotic-resistant pathogens in hospital wastewater, and these factors may spread into the receiving aquatic environment.
Similar content being viewed by others
Introduction
Antibiotics have been widely used to cure infectious diseases since they were discovered in the 1930s. In the environment, antibiotics are chemical pollutants that can exert toxic effects and are able to exert selection pressure on bacteria (González-Pleitera et al. 2019; Pazda et al. 2019). The excessive use and abuse of antibiotics have contributed immensely to the emergence of antibiotic-resistant bacteria (ARBs), such as vancomycin-resistant enterococci (Hocquet et al. 2016) and methicillin-resistant Staphylococcus aureus (Knight et al. 2012). Hospitals accumulate large amounts of antibiotics and human-related pathogens (Andersson and Hughes 2014). For example, Acinetobacter baumannii and Citrobacter freundii, which cause serious hospital-associated infections, have shown resistance to multiple antibiotics (Davies and Davies 2010). In addition, horizontal gene transfer of antibiotic resistance genes (ARGs) has aggravated the potential risk of antibiotic resistance evolution in recent years (Levy and Marshall 2004; Sorensen et al. 2005). Without suitable treatment, the discharge of ARBs and ARGs in hospital wastewater poses ecological and ARB evolution risks to aquatic environments and humans.
Large amounts of antibiotics are consumed in Chinese hospitals, especially β-lactams, quinolones, and trimethoprim (Boeckel et al. 2014; Zhang et al. 2015a; Klein et al. 2018). However, their occurrence in hospital wastewater is not yet understood. In recent years, studies have found several kinds of antibiotics (e.g. ofloxacin and cefalexin) in untreated hospital wastewater in the city of Xinxiang in central China (Wang et al. 2018). A similar result also showed that ofloxacin was detected at a high level in treated hospital wastewater (Rodriguez-Mozaz et al. 2015). In addition, most hospital wastewater is treated by independent hospital wastewater treatment plants (HWWTPs) and then discharged into the aquatic environment or downstream municipal wastewater treatment plants (MWWTPs). In recent years, many studies have focused on MWWTPs that receive wastewater from hospitals, communities, and industry (Lee et al. 2017; Quintela-Baluja et al. 2019). However, studies focusing on the hospital wastewater treatment process are limited, leaving a knowledge gap related to the occurrence of antibiotics, from traditional to last-line antibiotics, and their corresponding ARGs after hospital wastewater treatment protocols. Studying independent HWWTPs could help us understand the discharge of clinical antibiotics and ARGs and reveal the potential risks of hospital wastewater to the aquatic environment and receiving MWWTPs.
Hospitals can be divided into primary, secondary, and tertiary hospitals according to their medical capabilities and facilities (National Health Commission of the People’s Republic of China (NHCPRC) 1989; Jamison et al. 2006; Moore et al. 2014). Primary hospitals provide mostly basic health care for the community, while comprehensive and specialist health services are provided in tertiary hospitals (e.g. teaching hospitals, chest hospitals, and infectious disease hospitals) (World Health Organization (WHO) 1978; National Health Commission of the People’s Republic of China (NHCPRC) 1989). Differences in health services, bed capacity, antibiotic usage, amount of wastewater, and HWWTPs among hospitals of different scales may lead to different patterns of pollution of hospital wastewater with antibiotics, ARGs, and bacterial communities. In addition, antibiotics can sometimes be categorized into different generations according to their antimicrobial properties. For example, cephalosporins are grouped into the first, second, third, and fourth generations (Ribeiro et al. 2018). Revealing the contamination characteristics of different kinds of antibiotics and ARGs in different-scale hospitals could help us to understand the pollution status of antibiotics and the evolution of ARGs in hospital wastewater sources. To the best of our knowledge, this fundamental information has not yet been reported. Therefore, this study aims to (1) reveal the pollution characteristics of a variety of antibiotics and ARGs at different-scale hospitals in East China; (2) clarify the removal effectiveness of the conventional hospital wastewater treatment process for the target antibiotics and ARGs; and (3) evaluate the characteristics of the total microbial community and cultivable bacteria in untreated and treated hospital wastewater.
Materials and methods
Sample collection
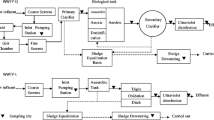
Samples were collected in triplicate in January and May 2019 from three different hospitals with different sizes and properties (primary, secondary, and tertiary hospitals, abbreviated H1, H2, and H3, respectively) in East China. These three hospitals had different independent treatment processes, with wastewater volumes of 80, 100, and 727 m3 per day. Briefly, H1 adopted a simple chlorine disinfection process because of its small scale and wastewater amount. At H2, the wastewater was first treated by a hydrolysis acidification tank and then by an aerobic contact tank, a secondary settling tank, and chlorine disinfection. H3 used an aerobic contact tank followed by a secondary settling tank and then chlorination treatment. The treated wastewater from these three hospitals was then discharged into receiving MWWTPs. Detailed information (e.g. hospital bed capacity, governance, and wastewater quality) of the three hospitals is listed in Table A.1. The samples were untreated wastewater and treated wastewater from the three HWWTPs, as shown in Fig. 1. The raw wastewater was a mixture of the medical ward and domestic wastewater. To avoid the possible degradation of β-lactam antibiotics, grab samples were collected in 5-L brown glass bottles on weekdays between 8:30 a.m. and 10:30 a.m., returned to the laboratory on ice in coolers, and analysed immediately. The containers were washed with methanol, water, and sterilized deionized water before wastewater collection. These included 18 samples for each season’s sampling campaign. Wastewater NH3-N and total phosphorus (TP) were analysed by a UV-Vis spectrophotometer (HACH DR 5000), and the chemical oxygen demand (COD) was measured by potassium dichromate titration, as shown in Table A.2. The concentrations of free chlorine were detected by the N,N-diethyl-p-phenylenediamine (DPD) colorimetric method (APHA 2017). The detailed chemicals and reagents that were used in this study are listed in Text A.1.
Quantification of antibiotics in hospital wastewater
Samples were analysed in triplicate for the determination of the target antibiotics following established methods (Gros et al. 2013; Cheng et al. 2015), with some modifications. Briefly, hospital wastewater was vacuum-filtered through a 0.7-μm glass fibre filter (Whatman GF/F, UK), followed by a 0.22-μm cellulose acetate membrane filter (Anpel, Shanghai, China). A suitable volume (1 mL) of a Na2EDTA solution (10 g/L) was added to the different types of water to achieve a final concentration of 0.1 g/L, and the pH of the samples was adjusted to 2.5 with hydrochloric acid. Water samples were automatically extracted using Oasis HLB cartridges (Wilford, MA, USA) for hospital wastewater matrices. SPE cartridges were activated with 6 mL of methanol followed by 6 mL of ultrapure water flowing at a gravity-dependent rate. Next, 100 mL of hospital wastewater was loaded onto the cartridge at a flow rate of 0.6–1 mL/min. After sample preconcentration, the cartridges were rinsed with 6 mL of HPLC grade water at a flow rate of 2 mL/min and vacuum-dried for 5 min to remove excess water. Following this, the cartridges were eluted with 10 mL of pure methanol at a flow rate of 1 ml/min. Then, the eluents were evaporated to approximately 100 μL under a gentle nitrogen stream and redissolved in 1 mL of methanol/water (1:1, v/v) solution.
The samples were finally analysed by liquid chromatography with tandem mass spectrometry (Shimadzu, LCMS-8050) using a Nexera X2 HPLC system equipped with a binary solvent manager and an autosampler, an electrospray ionization (ESI) interface, and LCMS LabSolutions software (Version 5.89). MS/MS parameters were optimized in multiple reaction monitoring (MRM) mode. An analytical Shim-pack GISS C18 column (2.1 × 100 mm, 1.9 μm, Shimadzu) was used at a flow rate of 0.4 mL/min, and the column oven temperature was set at 40°C. Mobile phase A (ultrapure water with 0.05% (v/v) formic acid) and mobile phase B (methanol) were used in stepwise gradient mode. The elution gradient started with 5% B, increased to 50% B over 4 min, then increased further to 80% B over 2 min, and finally increased to 95% B over 2 min; it was held at 95% B for 1 min and then brought back to the initial value over 0.1 min, where it was maintained for 2 min until the next injection. Mass spectra were acquired in positive ion mode with a nebulizer flow of 3 L/min, heating gas flow of 10 L/min, interface temperature of 250°C, desolvation line temperature of 150°C, heating block temperature of 400°C, and dry gas flow of 10 L/min.
To compare the characteristics of antibiotics in different-scale hospitals, typical antibiotics, such as first-, second-, third-, and fourth-generation cephalosporins, penicillins, carbapenem, quinolones, and trimethoprim, were selected. Standard first-generation cephalosporins (cefalexin (99%), cefalotin (99%), cefazolin (99%), cefradine (99%)), second-generation cephalosporins (cefoxitin (99%)), third-generation cephalosporins (ceftazidime (99%)), fourth-generation cephalosporins (cefepime (99%)), penicillins (amoxicillin (99%), ampicillin (99%), penicillin G (99%)), carbapenems (meropenem (99%)), quinolones (ofloxacin (99%), norfloxacin (99%)), and trimethoprim (99%) were purchased from Dr. Ehrenstorfer (Augsburg, Germany). For accurate quantification, stable isotope 13C3-caffeine (99%) (Augsburg, Germany) was spiked into the samples prior to SPE, and total recoveries were determined (n=3). The standard curve for each antibiotic was constructed with at least five concentrations (R2 > 0.99). The precursor ions, product ions, limits of detection (LODs), limits of quantification (LOQs), recoveries, linear R2 values, and reproducibility and repeatability results are shown in Table A.3. The low recovery values of β-lactam antibiotics may be attributed to their instability in hospital water matrices, and the detection of some antibiotic metabolites may benefit from the method used (Gros et al. 2013).
Characterization of bacteria and quantification of target genes
Hospital wastewater samples were serially diluted in 1× phosphate-buffered saline (PBS), and 100 μL volumes of samples were spread-plated in triplicate and incubated on lysogeny broth agar at 30°C for 48 h to determine the concentration of cultivable bacteria at the sampling site (Le et al. 2016). Colonies in the range of 30 to 300 cells were counted to minimize errors caused by the presence of too many colonies to count. One hundred millilitres of hospital wastewater was filtered through a 0.22-μm pore size cellulose nitrate membrane (Anpel, Shanghai, China). Total DNA was extracted from filters using the FastDNA SPIN Kit for Soil (MP Biomedicals, USA) according to the manufacturer’s instructions. DNA concentration and purity were measured using a NanoDrop spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific, Wilmington, DE, USA). The isolated DNA was stored at −20°C until subsequent analysis. Community characterization of total bacteria was performed by 16S amplicon sequencing at Sangon Biotech (Shanghai) Co., Ltd. (NCBI accession number: PRJNA646019). The V4 region of the 16S rRNA genes was amplified with the primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) and sequenced, and the diversity of the total bacteria was analysed. Specifically, the Illumina MiSeq raw sequencing data contained barcode, primer, and linker sequences. The primer and linker sequences were removed, and then the paired reads were merged into a sequence according to the relationship between paired-end reads. Samples were identified according to the individual barcodes to obtain data for each sample. The denoising process is described in the supporting information (Text A.2). Quality filtering of reads, trimming of paired ends, the denoising process, and the alpha diversity values from the bacterial diversity analysis are provided in the supplemental document (Table A.4, Table A.5, and Table A.6). In addition, for quantitative PCR (qPCR) analysis, DNA samples were diluted tenfold with nuclease-free water to minimize background interference in the qPCR reactions. qPCR was used to measure the abundances of 10 specific ARGs: the six β-lactam resistance genes blaOXA-1, blaOXA-10, blaTEM-1, blaDHA-1, blaSHV-1, and blaGES-1 and the four quinolone resistance genes qnrA, qnrS, qnrD, and qepA. The 16S rRNA gene and intI1 were analysed to determine the relative abundances of ARGs and the transferability of ARGs, respectively. All runs were performed with positive and negative controls. The positive controls were obtained by cloning target DNA into plasmids at different dilutions, and nuclease-free water was used as the negative control, as described in a previous study (Hu et al. 2019a). The details of the primers, annealing temperatures, and amplicons for the target genes were the same as those in our previous study and are listed in Table A.7 (Hu et al. 2019a). The detailed qPCR conditions were the same as those in our previous study and are listed in Text A.3 (Hu et al. 2019b).
Statistical analysis
The results are presented as the means ± standard deviations (SDs), and all diagrams were generated using R 3.6.1 (The R Foundation for Statistical Computing). The correlation analysis of antibiotics and ARGs was carried out using R 3.6.1 software. Correlation analysis of the target ARGs and mobile genetic elements was carried out using SPSS 25.0. Two-tailed P < 0.05 was considered to indicate significance.
Results and discussion
Analysis of hospital wastewater quality
The wastewater quality indices from the three hospitals are summarized in Table A.2. The treated wastewater quality indices met the active hospital wastewater pretreatment standard (Discharge Standard of Water Pollutants for Medical Organization, GB 18466-2005) (Table A.2). NH3-N, TP, and COD values decreased after HWWTP treatment. In particular, the NH3-N values ranged from 10.4 to 50 mg/L in the treated wastewater. NH3-N in wastewater can rapidly react with free chlorine to form chloramine, thereby reducing the concentration of free chlorine and the disinfection effect (Huang et al. 2012; Kevin 2016). Therefore, the breakpoint dosage (a mass ratio of chlorine to NH3-N of 7.6:1) of free chlorine was required in the presence of NH3-N in wastewater (Zhang et al. 2015b).
Occurrence of antibiotics in hospital wastewater
The pollution characteristics of antibiotics in hospital wastewater are shown in Fig. 2. The statistically significant differences in antibiotic pollution between the two sampling seasons were also analysed in Fig A.1, which revealed a nonsignificant difference. To provide a real picture of the pollution of antibiotics in hospital wastewater, the concentrations of the target antibiotics in the untreated wastewater and the removal efficiencies of the HWWTPs are summarized in Table 1. Thirteen antibiotics were detected in the wastewater of the three hospitals, but penicillin G was not detected. β-Lactams such as first-, second-, third-, and fourth-generation cephalosporins, penicillins, and carbapenem were widely detected in hospital wastewater. The diversity of antibiotics in the wastewater from the three hospitals varied greatly. For example, ofloxacin (2.38~9.23 μg/L) and the first-generation cephalosporin cefradine (0.37~2.38 μg/L) were detected at high concentrations in the wastewater from H1. The concentration of cefradine was higher than the concentration of cefradine (0.17 μg/L) reported in Taiwan, which may be attributed to its low clinical consumption (Li and Lin 2015). The main antibiotic types detected in the wastewater from H2 were quite different from those in the wastewater from H1. High concentrations of the second-generation cephalosporin cefoxitin (0.85~8.17 μg/L) and the third-generation cephalosporin ceftazidime (0.31~7.27 μg/L) were detected in wastewater from H2. The greatest diversity of antibiotics was detected in H3 compared with H1 and H2. H3 is the largest central hospital, receiving approximately 700,000 patients every year, and has independent intensive care units (ICUs). Thirteen kinds of antibiotics, including ten cephalosporins, two quinolones, and trimethoprim, were detected in wastewater from H3. Strikingly, the fourth-generation cephalosporin cefepime was detected only in wastewater from H3, with the highest concentration being 540.39 μg/L. The concentration of cefepime in this study was significantly higher than that reported in Romanian hospitals (8.52 μg/L). This might be explained by the smaller population served and the lower consumption of these antibiotics (providing service to 30,000 inhabitants) in the Romanian hospital (Szekeres et al. 2017). The carbapenem antibiotic meropenem, which is generally used as the last line of defence, was also detected in samples from H3 at a concentration of 0.20 μg/L. A higher concentration of meropenem (1.07 μg/L) was detected at a tertiary hospital that had 1500 beds (Le et al. 2016). Furthermore, the concentrations of the second-generation cephalosporin cefoxitin and the third-generation cephalosporin ceftazidime in wastewater from H3 were also higher than those in wastewater from H1 or H2. Notably, we found third-generation cephalosporin ceftazidime at all three hospitals; it is frequently used in Chinese hospitals according to the Status Report on Antimicrobial Administration and Antimicrobial Resistance in China, 2018 (National Health Commission of the People's Republic of China, www.nhc.gov.cn).
As shown in Fig. 2, ofloxacin was detected in all samples (including untreated and treated samples from the three hospitals) with high concentrations of 9.23 μg/L, 25.65 μg/L, and 49.47 μg/L in the samples from H1, H2, and H3, respectively. Similar results showed a high concentration of ofloxacin in a 360-bed hospital in the Essonne District, France (Dinh et al. 2017). However, the concentrations were higher than those in wastewater from Xinjiang Province, China, where the concentration of quinolones (e.g. ofloxacin, norfloxacin) ranged from 0.45 to 0.94 μg/L in untreated hospital wastewater (Li et al. 2016). Trimethoprim is often used with sulfonamide antibiotics to improve antibacterial properties. In this study, trimethoprim was detected at H1, H2, and H3 at concentrations of 0.02 μg/L, 0.31 μg/L, and 0.50 μg/L, respectively. The low concentrations of trimethoprim might be attributed to the small percentage of sulfonamides (e.g. sulfamethoxazole and sulfadiazine) that were used as human medicines in China. A recent study also showed that trimethoprim was detected at 0.78 to 0.84 μg/L in a hospital in Beijing, China (Liu et al. 2019). In recent years, total antibiotic consumption in many low- and middle-income countries was higher than that in high-income countries, with a rapidly increasing trend (Klein et al. 2018). The hospital consumption of antibiotics in China is the second largest in the world (Boeckel et al. 2014). Therefore, we suggest strengthening the management of antibiotics at hospitals and using antibiotics reasonably to reduce their discharge from hospital sources.
Removal of antibiotics at different HWWTPs
Considering that many kinds of antibiotics are widely detected in untreated hospital wastewater, the removal of antibiotics at different HWWTPs is further discussed. The removal rates of the target antibiotics at different HWWTPs ranged from 20.2 to 100% (Table 1). Specifically, the removal rates of β-lactams (including cephalosporins, penicillins, and carbapenem) reached 100% at the independent HWWTPs H1 and H3. Similar results showed high removal efficiencies of β-lactam antibiotics in biological wastewater treatment processes (84.4–99.5%) (Tran et al. 2016). It has been reported that the removal efficiency of antibiotics in hospital wastewater treatment processes ranges from −74.0 to 81.0% (Szekeres et al. 2017). High concentrations of β-lactams remained in the treated wastewater from H2, with the highest concentration being 2.95 μg/L. This differential result may be attributed to the fact that the removal of β-lactams is strongly affected by chemical or hydraulic retention times (Hou and Poole 1971; Le-Minh et al. 2010). The concentration of chlorine at H2 was relatively lower than those at H1 and H3 (Table A.2). It is also generally observed that increasing the removal efficiency of β-lactams demands more oxidizing agents (chlorine) in natural water matrices (Acero et al. 2010). The third-generation cephalosporin ceftazidime exhibited a removal efficiency of 51.4%. The release of critical β-lactam antibiotics (ceftazidime) may enhance antibiotic resistance in aquatic environments.
Ofloxacin was recalcitrant and detected with a frequency of almost 100% in the treated effluents from the three hospitals. Many studies have reported that ofloxacin is detected more frequently than other antibiotics in hospital wastewater (Rodriguez-Mozaz et al. 2015; Wang et al. 2018). The removal efficiencies of ofloxacin at the HWWTPs of H1 and H2 were 44.2% and 51.5%, respectively. Although the removal efficiency of ofloxacin at the HWWTP of H3 was improved (68.8%), this antibiotic still presented a high concentration of 14.90 μg/L in the treated wastewater. This result was consistent with those of previous studies that proved that traditional hospital wastewater treatment systems applying biological processes, biological anoxic reactors, and membrane filtration methods exhibited limited removal efficiency for ofloxacin (Kovalova et al. 2012; Dinh et al. 2017). The low removal efficiency of ofloxacin may be attributed to the chemically stable piperazinyl ring and its lack of biodegradation vulnerabilities (Turiel et al. 2005; Hapeshi et al. 2013). Considering that the conventional hospital treatment process was not designed to remove antibiotics, emerging powerful options for the treatment of antibiotics have been studied in recent years. For example, quinolones can be efficiently removed by simulated solar radiation systems (Babic et al. 2013). The emerging electro-Fenton process has shown good removal of antibiotics and has the advantages of low cost and reusable solid catalysts as sources of Fe2+ and Cu2+ ions (Barhoumi et al. 2016; Barhoumi et al. 2017).
Occurrence of ARGs in hospital wastewater
The prevalence of ARGs (normalized to 16S rRNA and log transformed) in hospital wastewater is shown in Fig. 3. The relative abundances of the analysed ARGs are listed in Table A.8. Fig A.2 shows the statistically significant differences in ARGs pollution between the two sampling seasons, and a nonsignificant difference was found. The order of the average relative abundances of target ARGs and intI1 was as follows: H3 (1.35×10−4 copies/16S rRNA gene) > H2 (1.30×10−4 copies/16S rRNA gene) > H1 (6.25×10−5 copies/16S rRNA gene) (Fig. 3). Therefore, the pollution of ARGs at tertiary hospital H3 was more serious than that at primary hospital H1 and secondary hospital H2. The most abundant ARGs in the wastewater from H1, H2, and H3 were blaTEM-1, blaGES-1, and blaOXA-1, respectively. The relative abundances of the carbapenem resistance gene blaGES-1 in the wastewater from H1, H2, and H3 were 6.21×10−5, 1.77×10−3, and 9.44×10−4 copies/16S rRNA gene, respectively. In addition, the cephalosporin resistance gene blaOXA-1 showed a high relative abundance of 1.27×10−3 copies/16S rRNA gene at three hospitals. blaOXA-1 is frequently detected (57.9%) in Escherichia coli resistant to cefotaxime in hospital wastewater (Adegoke et al. 2020). Mobile genetic elements or plasmid-associated OXA β-lactamase genes were found to be significantly enriched in the plasmid reads of secondary wastewater samples (Barlow and Hall 2002; Martin et al. 2020). To provide a real picture of ARG pollution in wastewater, the ARG pollution in this study and data from other parts of the world is summarized in Table A.9. A previous systematic review also found the presence of antibiotics (e.g. β-lactam, quinolone, trimethoprim, and multidrug efflux genes) in municipal, hospital, and industrial wastewater in different countries (e.g. USA, Canada, and China) (Pazda et al. 2019). The class 1 integron displayed a relative abundance of 1.17×10−4 copies/16S rRNA gene in untreated hospital wastewater. The class 1 integron is a representative mobile gene element that can sometimes enhance the HGT of ARGs. The removal rate for the class 1 integron at H3 was significantly higher than those at H1 and H2 (Fig. 4). In previous work, less than 1.2 log of intI1 was removed by hospital wastewater treatment, and intI1 remained detectable at 8×10−5 copies/16S rRNA gene (Timraz et al. 2017). Forty-three ARG cassettes mediated by intI1 were detected with great diversity in hospital wastewater (Yuan et al. 2021). As indicated in Table A.10, the presence of the last-line antibiotic (carbapenem) resistance gene blaGES-1 was strongly associated with the presence of blaOXA-10. In addition, the presence of blaOXA-10, blaDHA-1, blaSHV-1, qnrA, and qnrS was significantly correlated with the presence of intI1 at the 0.01 level. This phenomenon may be attributed to the presence of these genes in the same gene cassettes that could enhance the risk of ARG transfer, which was also proven by some studies (Ma et al. 2017; An et al. 2018).
Quinolone ARGs decreased in abundance in the order qnrA > qnrS > qnrD > qeqA, which was consistent with the results of a previous study showing that qnrA was the quinolone ARG with the highest concentration (Wang et al. 2018). The concentration of qnrA (1.44×105 copies/mL) in this study was one magnitude higher than that in the above study (1.58×104 copies/mL). In addition, the average concentration of β-lactam resistance genes (4.87×106 copies/mL) was two orders of magnitude higher than that of quinolone resistance genes (5.38×104 copies/mL), which were present at 1.53×10−6 copies/16S rRNA gene in treated wastewater from the three hospitals. Therefore, these findings suggested that the risk of β-lactam antibiotics and the related ARGs should be given more attention and considered to improve the clearance efficiency for ARGs in HWWTPs that have the possibility of harbouring the targeted ARGs (Ju et al. 2016).
In addition, correlations of all the targeted types of antibiotics with their corresponding ARGs were further evaluated (Fig. A.3). A significantly positive correlation was revealed between blaGES-1 and the total β-lactam concentration (P < 0.01) and between blaOXA-1 and the total β-lactam concentration (P < 0.05). This result suggested that the high concentration and variety of β-lactams may be responsible for the high concentrations of β-lactam ARGs. However, blaSHV-1, blaTEM-1, and qnrD had weak correlations with the corresponding antibiotics. A strong correlation between qnrA and the total quinolone concentration (P < 0.05) and between qnrS and the total quinolone concentration (P < 0.05) was observed. A previous study showed a significant correlation between the total concentration of plasmid-mediated quinolone resistance genes and the corresponding antibiotics in wastewater and soil samples (Li et al. 2012). Similarly, strong correlations between the qnrS gene and ofloxacin and ciprofloxacin were observed (Rodriguez-Mozaz et al. 2015). Exposure to antibiotics could exert selective pressure on bacteria and increase the concentration of ARGs in the environment. Although we have demonstrated a significant correlation between several types of antibiotics and ARGs, the environmental influencing factors and other pollutants in the hospital wastewater matrix need to be further evaluated to provide a better understanding of the co-occurrence of these pollutants.
Removal of ARGs at different HWWTPs
The removal efficiencies of ARGs at different HWWTPs are shown in Fig. 4. The removal rates of the target ARGs at the three HWWTPs ranged from −0.85 to 2.71 log in both sampling campaigns (Fig. 4). Specifically, the log reductions were the highest for qnrS, blaSHV-1, and blaDHA-1 among the target ARGs. Similar to the results of a previous study, qnrS and blaSHV were reduced by approximately one order of magnitude at traditional wastewater treatment plants (Laht et al. 2014; Rodriguez-Mozaz et al. 2015). The traditional wastewater treatment process cannot efficiently remove target ARGs from hospital wastewater. Notably, the abundances of five β-lactam ARGs (blaOXA-1, blaOXA-10, blaDHA-1, blaSHV-1, and blaTEM-1) and two quinolone ARGs (qnrA and qnrD) increased after treatment at H1 in Jan. Several studies have proven that the chlorination process can increase ARG concentrations. For example, the levels of aminoglycoside-resistant genes (aac(6')-II and aacC2) and tetracycline-resistance genes (tetR and tetX) increased from 0.22 to 2.23 log (Wang et al. 2020). Several ARGs (e.g. qnrB, tetM, and tetW) were also increased after disinfection with sodium hypochlorite (Hu et al. 2019a). The increases in ARGs may be attributed to an inadequate chlorine dosage, inducing the formation of more pili for conjugative transfer (Guo et al. 2015). The horizontal transfer of ARGs was easily induced at a nonlethal dose of chlorine (0.5 mg/L) (Wang et al. 2020). Therefore, the enhanced disinfection efficiency and the inhibition of ARG transferability in hospital wastewater need to be considered. In the present study, the relative abundances of ARGs were between 1.01×10−9 and 1.77×10−3 copies/16S rRNA gene in the treated wastewater. The residual ARBs and ARGs in the treated wastewater are further sent to MWWTPs and become potential sources of gene propagation and horizontal transfer (Xu et al. 2015; Bengtsson-Palme et al. 2016). These results suggested that the HWWTPs had a poor removal efficiency for ARGs and that it is necessary to improve hospital wastewater treatment facilities to remove these ARGs and mobile genetic elements. In recent years, potential methods that have shown good removal ability for these novel pollutants, such as bioelectroreduction, UV/chlorination, and radiation, have been applied in hospital wastewater treatment after careful testing (Liang et al. 2019; Zhang et al. 2019).
Analysis of bacteria and community characterization in hospital wastewater samples
Figure 5 shows the results of bacterial abundance and community characterization in the untreated and treated wastewater of the three hospitals considered in this work, namely H1, H2, and H3. In Fig. 5a, abundance values are expressed as the 16S rRNA gene copy number per millilitre, and as shown, the concentration of the 16S rRNA gene ranged from 2.93×109 to 8.85×1010 copies/mL in the wastewater, which were higher than the values reported in a previous study (Szekeres et al. 2017). This may be attributed to the smaller number of people served (around 30,000 inhabitants) in Cluj County, Romania (Szekeres et al. 2017). The removal rates of the 16S rRNA gene showed a declining trend, with removal ranging from 0.05 to 1.24 log. The number of culturable bacteria in the untreated wastewater increased in the order H1 < H2 < H3 (Fig. 5b), while it increased in the order H3 < H1 <H2 in the treated wastewater, which may be explained by the low total chlorine level at H2 (Table A.2). The culturable bacterial concentration ranged from 2.0×104 to 2.0×105 CFU/mL in the treated hospital wastewater. A similar result showed that live bacteria were still present at a density of 2.6×104 CFU/mL in the treated wastewater (Lee et al. 2017). Ceftazidime-resistant bacteria ranged from 104 to 106 CFU/mL at two hospitals in Singapore (Le et al. 2016). β-Lactam-resistant bacteria can destroy even novel penicillins and cephalosporins (Levy and Marshall 2004). It has been reported that chlorine disinfection cannot completely inactivate ceftazidime-resistant bacteria, and there remains a potential risk of releasing the bacteria to the environment (Beattie et al. 2020). These results indicated that the treated hospital wastewater still contained high concentrations of microorganisms, especially live bacteria. These surviving bacteria may further proliferate and pose a risk for receiving MWWTPs.
The communities in the untreated and treated H1, H2, and H3 hospitals were also analysed. The bacterial compositions in hospital wastewater differed at the genus level (Fig. 5c). For example, sequences affiliated with the genera Aquitalea, Thauera, and Romboutsia were detected at the highest abundance in the untreated wastewater from H1, H2, and H3, respectively. Notably, several genera related to (opportunistic) pathogens, such as Acinetobacter (3.59%), Klebsiella (2.07%), Aeromonas (8.84%), and Pseudomonas (7.60%), were found at relatively high abundances in hospital wastewater. This was similar to the results of a previous study, which showed that Acinetobacter and Aeromonas were the dominant (opportunistic) pathogens in untreated hospital water (Wang et al. 2018). Acinetobacter is a leading emerging opportunistic pathogen that is frequently reported at hospitals. For example, multiple antibiotic-resistant forms of Acinetobacter baumannii were discharged from hospital sites and hospital wastewater into receiving urban wastewater (Kovacic et al. 2017; Music et al. 2017). Aeromonas species showed resistance not only to most cephalosporins and penicillins but also to carbapenems, which are usually used as the last reliable antibiotic treatment. In recent years, the opportunistic pathogen Aeromonas sp. has frequently been reported to contain both genomic and plasmid-mediated carbapenem resistance genes (Mathys et al. 2019).
In addition, the sequences affiliated with the genera Pseudomonas and Spirillum were dominant in treated wastewater from all three hospitals, accounting for 4.37–16.94% of the total genera. Pseudomonas aeruginosa is a common opportunistic human bacterium that is frequently detected in hospital wastewater; it has been reported to convey resistance to third- and fourth-generation cephalosporins and carbapenems and to persist after treatment with oxidizing chemicals (Polotto et al. 2012; Hou et al. 2019). Another typical human bacterium, Escherichia coli, which is strongly related to some human diseases, was also present in the treated wastewater from primary hospital H1. Mycobacterium is also a typical human pathogen with a unique cell wall that survives under long-term exposure to many antibiotics, oxidants, and even chlorine, and it presents higher relative abundances in the treated wastewater from H2 and H3 than in that from H1. Pathogens from hospital wards, ICUs, and toilets are discharged into HWWTPs, where the pathogens may be further propagated and extensively spread. A recent study showed that the abundance of multi-ARBs in hospital wastewater was significantly greater than that in non-hospital wastewater (Moges et al. 2014). Furthermore, it has been shown that hospitals are the main source of carbapenem-resistant pathogens in the environment (Lamba et al. 2017).
In addition, the presence of long-term chlorine disinfectants makes it possible for microorganisms to develop chlorine resistance (Luo et al. 2021). Considering that hospital wastewater contains a variety of clinically related antibiotics and pathogens, pathogens that show both antibiotic and chlorine resistance may enhance the potential risk of transmission to the aquatic environment and humans. Given the severe impact of the outbreak of COVID-19 in 2020, hospital wastewater also became a potential reservoir of the virus (Saguti et al. 2020). As discussed in the present study, the occurrence of antibiotics and the incomplete disinfection of ARGs and ARBs in HWWTPs indicate the potential release of these pollutants. Therefore, novel effective disinfection techniques for ARBs, ARGs, or even viruses in hospital wastewater matrices warrant further attention.
Conclusion
This study investigated the occurrence and removal of antibiotics, their corresponding ARGs, and human-related pathogens in different-scale hospitals. The occurrence of antibiotics and ARGs in wastewater from three hospitals varied greatly. Wastewater from primary hospital H1 frequently contained traditional antibiotics (e.g. ofloxacin and cefradine), while wastewater from tertiary hospital H3 contained new antibiotics (e.g. the fourth-generation cephalosporin cefepime and the carbapenem meropenem). HWWTPs were not efficient at removing all ARGs, and the concentrations of several ARGs were increased in the treated wastewater. Antibiotics and ARGs have the potential risk of spreading into downstream MWWTPs and aquatic environments from hospital wastewater. The treated hospital wastewater still contained high concentrations of microorganisms, especially live bacteria. Several opportunistic or pathogenic bacteria (e.g. Acinetobacter, Klebsiella, Aeromonas, and Pseudomonas) had high proportions in the treated wastewater. These results provide the basis for the co-occurrence of new antibiotics, ARGs, and antibiotic-resistant pathogens in hospital sources and highlight the need to eliminate these contaminants in the hospital source.
Data Availability
The availability of data and materials is on the base of personal request.
References
Acero JL, Benitez FJ, Real FJ, Roldan G (2010) Kinetics of aqueous chlorination of some pharmaceuticals and their elimination from water matrices. Water Res 44:4158–4170
Adegoke AA, Madu CE, Aiyegoro OA, Stenstrom TA, Okoh AI (2020) Antibiogram and beta-lactamase genes among cefotaxime resistant E. coli from wastewater treatment plant. Antimicrob. Resist Infect Control 9:46–57
An XL, Chen QL, Zhu D, Zhu YG, Gillings MR, Su JQ (2018) Impact of wastewater treatment on the prevalence of integrons and the genetic diversity of integron gene cassettes. Appl Environ Microbiol 84:e02766–e02717
Andersson DI, Hughes D (2014) Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478
APHA (2017) Standard methods for the examination of water and wastewater. American. Public Health Association, American Water Works Association, Water Environment Federation, Washington DC
Babic S, Perisa M, Skoric I (2013) Photolytic degradation of norfloxacin, enrofloxacin and. ciprofloxacin in various aqueous media. Chemosphere 91:1635–1642
Barhoumi N, Oturan N, Olvera-Vargas H, Brillas E, Gadri A, Ammar S, Oturan MA (2016) Pyrite as a sustainable catalyst in electro-Fenton process for improving oxidation of sulfamethazine. Kinetics, mechanism and toxicityassessment. Water Res 94:52–61
Barhoumi N, Olvera-Vargas H, Oturan N, Huguenot D, Gadri A, Ammar S, Brillas E, Oturan MA (2017) Kinetics of oxidative degradation/mineralization pathways of the antibiotic tetracycline by the novel heterogeneous electro-Fenton process with solid catalyst chalcopyrite. Appl Catal B-Environ 209:637–647
Barlow M, Hall BG (2002) Phylogenetic analysis shows that the OXA beta-lactamase genes have been on plasmids for millions of years. J Mol Evol 55:314–321
Beattie R, Skwor T, Hristova K (2020) Survivor microbial populations in post-chlorinated wastewater are strongly associated with untreated hospital sewage and include ceftazidime and meropenem resistant populations. Sci Total Environ 740:140186
Bengtsson-Palme J, Hammaren R, Pal C, Ostman M, Bjorlenius B, Flach CF, Fick J, Kristiansson E, Tysklind M, Larsson DGJ (2016) Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci Total Environ 572:697–712
Boeckel TPV, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, Laxminarayan R (2014) Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 14:742–750
Cheng WH, Jiang L, Lu N, Ma L, Sun XY, Luo Y, Lin KF, Cui CZ (2015) Development of a method for trace level determination of antibiotics in drinking water sources by high performance liquid chromatography-tandem mass spectrometry. Anal Methods 7:1777–1787
Davies J, Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433
Dinh QT, Moreau-Guigon E, Labadie P, Alliot F, Teil M, Blanchard M, Eurin J, Chevreuil M (2017) Fate of antibiotics from hospital and domestic sources in a sewage network. Sci Total Environ 575:758–766
González-Pleitera M, Cirésa S, Hurtado-Gallego J, Leganés F, Fernández-Piñas F, Velázquez D (2019) Cyanobacteria: from basic science to applications. In: Chapter 20 - Ecotoxicological assessment of antibiotics in freshwater using Cyanobacteria. Academic Press, New York, pp 399–417
Gros M, Rodriguez-Mozaz S, Barcelo D (2013) Rapid analysis of multiclass antibiotic residues and some of their metabolites in hospital, urban wastewater and river water by ultra-high-performance liquid chromatography coupled to quadrupole-linear ion trap tandem mass spectrometry. J Chromatogr A 1292:173–188
Guo MT, Yuan QB, Yang J (2015) Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater. Environ Sci Technol 49:5771–5778
Hapeshi E, Fotiou I, Fatta-Kassinos D (2013) Sonophotocatalytic treatment of ofloxacin in secondary treated effluent and elucidation of its transformation products. Chem Eng J 224:96–105
Hocquet D, Muller A, Bertrand X (2016) What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect 93:395–402
Hou JP, Poole JW (1971) β-Lactam antibiotics: their physicochemical properties and biological activities in relation to structure. J Pharm Sci 60:503–532
Hou AM, Yang D, Miao J, Shi DY, Yin J, Yang ZW, Shen ZQ, Wang HR, Qiu ZJ, Liu WL, Li JW, Li JW, Jin M (2019) Chlorine injury enhances antibiotic resistance in Pseudomonas aeruginosa through over expression of drug efflux pumps. Water Res 156:366–371
Hu YR, Zhang TY, Jiang L, Luo Y, Yao SJ, Zhang D, Lin KF, Cui CZ (2019a) Occurrence and reduction of antibiotic resistance genes in conventional and advanced drinking water treatment processes. Sci Total Environ 669:777–784
Hu YR, Zhang TY, Jiang L, Yao SJ, Ye H, Lin KF, Cui CZ (2019b) Removal of sulfonamide antibiotic resistant bacterial and intracellular antibiotic resistance genes by UVC-activated peroxymonosulfate. Chem Eng J 368:888–895
Huang HM, Xiao XM, Yan B (2012) Complex treatment of the ammonium nitrogen wastewater from rare-earth separation plant. Desalin Water Treat 8:109–117
Jamison DT, Breman JG, Measham AR, Alleyne G, Claeson M, Evans DB, Jha P, Mills A, Musgrove P (2006) Disease control priorities in developing countries, second edn. Oxford University Press, Washington DC
Ju F, Li B, Ma LP, Wang YB, Huang DP, Zhang T (2016) Antibiotic resistance genes and human bacterial pathogens: co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res 91:1–10
Kevin H (2016) Effects of chlorination on the inactivation and reactivation of Escherichia coli K12 and its ampicillin resistance gene. Dissertation, University of California
Klein EY, Boeckel TPV, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R (2018) Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A 115:E3463–E3470
Knight GM, Budd EL, Whitney L, Thornley A, Al-Ghusein H, Planche T, Lindsay JA (2012) Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. J Antimicrob Chemother 67:2514–2522
Kovacic A, Music MS, Dekic S, Tonkic M, Novak A, Rubic Z, Hrenovic J, Goic-Barisic I (2017) Transmission and survival of carbapenem-resistant Acinetobacter baumannii outside hospital setting. Int Microbiol 20:165–169
Kovalova L, Siegrist H, Singer H, Wittmer A, McArdell CS (2012) Hospital wastewater treatment by membrane bioreactor: performance and efficiency for organic micropollutant elimination. Environ Sci Technol 46:1536–1545
Laht M, Karkman A, Voolaid V, Ritz C, Tenson T, Virta M, Kisand V (2014) Abundances of. tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS One 9:e103705
Lamba M, Graham DW, Ahammad SZ (2017) Hospital wastewater releases of carbapenem-resistance pathogens and genes in Urban India. Environ Sci Technol 51:13906–13912
Le TH, Ng C, Chen HJ, Yi XZ, Koh TH, Barkham TMS, Zhou Z, Gina KYH (2016) Occurrences and characterization of antibiotic-resistant bacteria and genetic determinants of hospital wastewater in a tropical country. Antimicrob Agents Chemother 60:7449–7456
Lee J, Jeon JH, Shin J, Jang HM, Kim S, Song MS, Kim YM (2017) Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. Sci Total Environ 605-606:906–914
Le-Minh N, Khan SJ, Drewes JE, Stuetz RM (2010) Fate of antibiotics during municipal water. recycling treatment processes. Water Res 44:4295–4323
Levy SB, Marshall B (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10:S122–S129
Li SW, Lin AY (2015) Increased acute toxicity to fish caused by pharmaceuticals in hospital effluents in a pharmaceutical mixture and after solar irradiation. Chemosphere 139:190–196
Li J, Wang T, Shao B, Shen JZ, Wang SC, Wu YN (2012) Plasmid-mediated quinolone resistance genes and antibiotic residues in wastewater and soil adjacent to swine feedlots: potential transfer to agricultural lands. Environ Health Perspect 120:1144–1149
Li C, Lu JJ, Liu J, Zhang GL, Tong YB, Ma N (2016) Exploring the correlations between antibiotics and antibiotic resistance genes in the wastewater treatment plants of hospitals in Xinjiang, China. Environ Sci Pollut Res Int 23:15111–15121
Liang B, Ma JC, Cai WW, Li Z, Liu WZ, Qi MY, Zhao YK, Ma XD, Deng Y, Wang AJ, Zhou JZ (2019) Response of chloramphenicol-reducing biocathode resistome to continuous electrical stimulation. Water Res 148:398–406
Liu XH, Zhang GD, Liu Y, Lu SY, Qin P, Guo XC, Bi B, Wang L, Xi BD, Wu FC, Wang WL, Zhang TT (2019) Occurrence and fate of antibiotics and antibiotic resistance genes in typical urban water of Beijing, China. Environ Pollut 246:163–173
Luo LW, Wu YH, Yu T, Wang YH, Chen GQ, Tong X, Bai Y, Xu C, Wang HB, Ikuno N, Hu HY (2021) Evaluating method and potential risks of chlorine-resistant bacteria (CRB): a review. Water Res 188:116474
Ma LP, Li AD, Yin XL, Zhang T (2017) The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments. Environ Sci Technol 51:5721–5728
Martin C, Stebbins B, Ajmani A, Comendul A, Hamner S, Hasan NA, Rita Colwell R, Timothy FT (2020) Nanopore-based metagenomics analysis reveals prevalence of mobile antibiotic and heavy metal resistome in wastewater. Ecotoxicology.
Mathys DA, Mollenkopf DF, Feicht SM, Adams RJ, Albers AL, Stuever DM, Grooters SV, Ballash GA, Daniels JB, Wittum TE (2019) Carbapenemase-producing Enterobacteriaceae and Aeromonas spp. present in wastewater treatment plant effluent and nearby surface waters in the US. PLoS One 14:e0218650
Moges F, Endris M, Belyhun Y, Worku W (2014) Isolation and. characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC Res Notes 7:215–220
Moore LSP, Freeman R, Gilchrist MJ, Gharbi M, Brannigan ET, Donaldson H, Livermore DM, Holmes AH (2014) Homogeneity of antimicrobial policy, yet heterogeneity of antimicrobial resistance: antimicrobial non-susceptibility among 108717 clinical isolates from primary, secondary and tertiary care patients in London. J Antimicrob Chemother 69:3409–3422
Music SM, Hrenovic J, Goic-Barisic I, Hunjak B, Skoric D, Ivankovic T (2017) Emission of extensively-drug-resistant Acinetobacter baumannii from hospital settings to the natural environment. J Hosp Infect 96:323–327
National Health Commission of the People’s Republic of China (NHCPRC) (1989). Administrative measures for hospital classification. National Health Commission of the People’s Republic of China.
Pazda M, Kumirska J, Stepnowski P, Mulkiewicz E (2019) Antibiotic resistance genes identified in wastewater treatment plant systems - a review. Sci Total Environ 697:134023
Polotto M, Casella T, Oliveira MGL, Rúbio FG, Nogueira ML, Almeida MT, Nogueira MC (2012) Detection of P. aeruginosa harboring blaCTX-M-2, blaGES-1 and blaGES-5, blaIMP-1 and blaSPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis 12:176–183
Quintela-Baluja M, Abouelnaga M, Romalde J, Su JQ, Yu YJ, Gomez-Lopez M, Smets B, Zhu YG, Graham DW (2019) Spatial ecology of a wastewater network defines the antibiotic resistance genes in downstream receiving waters. Water Res 162:347–357
Ribeiro AR, Sures B, Schmidt TC (2018) Cephalosporin antibiotics in the aquatic environment: a critical review of occurrence, fate, ecotoxicity and removal technologies. Environ Pollut 241:1153–1166
Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sanchez-Melsio A, Borrego CM, Barcelo D, Balcazar JL (2015) Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res 69:234–242
Saguti F, Magnil E, Enache L (2020) Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res 189:116620
Sorensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S (2005) Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Microbiol 3:700–710
Szekeres E, Baricz A, Chiriac CM, Farkas A, Opris O, Soran M, Andrei A, Rudi K, Balcazar JL, Dragos N, Coman C (2017) Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ Pollut 225:304–315
Timraz K, Xiong YH, Qarni HA, Hong PY (2017) Removal of bacterial cells, antibiotic resistance genes and integrase genes by on-site hospital wastewater treatment plants: surveillance of treated hospital effluent quality. Environ Sci Water Res Technol 3:293–303
Tran NH, Chen HJ, Reinhard M, Mao FJ, Gin KY (2016) Occurrence and removal of multiple classes of antibiotics and antimicrobial agents in biological wastewater treatment processes. Water Res 104:461–472
Turiel E, Bordin G, Rodriguez AR (2005) Study of the evolution and degradation products of ciprofloxacin and oxolinic acid in river water samples by HPLC-UV/MS/MS-MS. J Environ Monit 7:189–195
Wang Q, Wang PL, Yang QX (2018) Occurrence and diversity of antibiotic resistance in untreated hospital wastewater. Sci Total Environ 621:990–999
Wang HC, Wang J, Li SM, Ding GY, Wang K, Zhuang T, Huang X, Wang XY (2020) Synergistic effect of UV/chlorine in bacterial inactivation, resistance gene removal, and gene conjugative transfer blocking. Water Res 185:116290
World Health Organization (WHO) (1978). Primary health care. World Health Organization.
Xu J, Xu Y, Wang HM, Guo CS, Qiu HY, He Y, Zhang Y, Li XC, Meng W (2015) Occurrence of. antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 119:1379–1385
Yuan W, Zhang YL, Riaz L, Yang QX, Du BB, Wang RF (2021) Multiple antibiotic resistance and DNA methylation in Enterobacteriaceae isolates from different environments. J Hazard Mater 402:123822
Zhang QQ, Ying GG, Pan CG, Liu YS, Zhao JL (2015a) Comprehensive evaluation of. antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ Sci Technol 49:6772–6782
Zhang YY, Zhuang Y, Geng JJ, Ren HQ, Zhang Y, Ding LL, Xu K (2015b) Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci Total Environ 512-513:125–132
Zhang TY, Hu YR, Jiang L, Yao SJ, Lin KF, Zhou YB, Cui CZ (2019) Removal of antibiotic resistance genes and control of horizontal transfer risk by UV, chlorination and UV/chlorination treatments of drinking water. Chem Eng J 358:589–597
Funding
This work was supported by the Science and Technology Commission of Shanghai Municipality (No. 20dz1200800) and China National Major Science and Technology Program for Water Pollution Control and Treatment (No. 2017ZX07402003).
Author information
Authors and Affiliations
Contributions
The manuscript was reviewed and approved for publication by all authors. CC and SY conceived and designed the experiments. SY and QY performed the experiments. YH and JY analysed the data. SY and CC wrote the paper. SY, JY, QY, YH, TZ, LJ, SM, KL, and CC reviewed and revised the paper.
Corresponding author
Ethics declarations
Ethical approval
The manuscript was reviewed and ethical approved for publication by all authors.
Consent to participate
The manuscript was reviewed and consents to participate by all authors.
Consent to publish
The manuscript was reviewed and consents to publish by all authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Ester Heath
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 99374 kb)
Rights and permissions
About this article
Cite this article
Yao, S., Ye, J., Yang, Q. et al. Occurrence and removal of antibiotics, antibiotic resistance genes, and bacterial communities in hospital wastewater. Environ Sci Pollut Res 28, 57321–57333 (2021). https://doi.org/10.1007/s11356-021-14735-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-021-14735-3