Abstract
Purpose
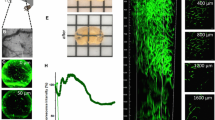
The goals of this study were to create cryo-imaging methods to quantify characteristics (size, dispersal, and blood vessel density) of mouse orthotopic models of glioblastoma multiforme (GBM) and to enable studies of tumor biology, targeted imaging agents, and theranostic nanoparticles.
Procedures
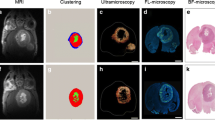
Green fluorescent protein-labeled, human glioma LN-229 cells were implanted into mouse brain. At 20–38 days, cryo-imaging gave whole brain, 4-GB, 3D microscopic images of bright field anatomy, including vasculature, and fluorescent tumor. Image analysis/visualization methods were developed.
Results
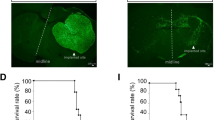
Vessel visualization and segmentation methods successfully enabled analyses. The main tumor mass volume, the number of dispersed clusters, the number of cells/cluster, and the percent dispersed volume all increase with age of the tumor. Histograms of dispersal distance give a mean and median of 63 and 56 μm, respectively, averaged over all brains. Dispersal distance tends to increase with age of the tumors. Dispersal tends to occur along blood vessels. Blood vessel density did not appear to increase in and around the tumor with this cell line.
Conclusion
Cryo-imaging and software allow, for the first time, 3D, whole brain, microscopic characterization of a tumor from a particular cell line. LN-229 exhibits considerable dispersal along blood vessels, a characteristic of human tumors that limits treatment success.









Similar content being viewed by others
References
Iacob G, Dinca EB (2009) Current data and strategy in glioblastoma multiforme. J Med Life 2:386–393
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21:2683–2710
Ichimura K, Ohgaki H, Kleihues P, Collins VP (2004) Molecular pathogenesis of astrocytic tumours. J Neurooncol 70:137–160
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114:97–109
Louis DN (2006) Molecular pathology of malignant gliomas. Annu Rev Pathol 1:97–117
Teodorczyk M, Martin-Villalba A (2010) Sensing invasion: cell surface receptors driving spreading of glioblastoma. J Cell Physiol 222:1–10
Candolfi M, Curtin JF, Nichols WS, Muhammad AG, King GD, Pluhar GE, McNiel EA, Ohlfest JR, Freese AB, Moore PF, Lerner J, Lowenstein PR, Castro MG (2007) Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression. J Neurooncol 85:133–148
Barth RF, Kaur B (2009) Rat brain tumor models in experimental neuro-oncology: the C6, 9L, T9, RG2, F98, BT4C, RT-2 and CNS-1 gliomas. J Neurooncol 94:299–312
Gargesha M, Qutaish MQ, Roy D, Steyer GJ, Watanabe M, Wilson DL (2011) Visualization of color anatomy and molecular fluorescence in whole-mouse cryo-imaging. Comput Med Imaging Graph 35:195–205
Krishnamurthi G, Wang CY, Steyer G, Wilson DL (2010) Removal of subsurface fluorescence in cryo-imaging using deconvolution. Opt Express 18:22324–22338
Roy D, Steyer GJ, Gargesha M, Stone ME, Wilson DL (2009) 3D cryo-imaging: a very high-resolution view of the whole mouse. Anat Rec (Hoboken) 292:342–351
Steyer GJ, Roy D, Salvado O, Stone ME, Wilson DL (2009) Removal of out-of-plane fluorescence for single cell visualization and quantification in cryo-imaging. Ann Biomed Eng 37:1613–1628
Wilson D, Roy D, Steyer G, Gargesha M, Stone M, McKinley E (2008) Whole mouse cryo-imaging. Proc Soc Photo Opt Instrum Eng 6916:69161I–69161I9
Burden-Gulley SM, Gates TJ, Burgoyne AM, Cutter JL, Lodowski DT, Robinson S, Sloan AE, Miller RH, Basilion JP, Brady-Kalnay SM (2010) A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase mu. Neoplasia 12:305–316
Zou KH, Warfield SK, Bharatha A, Tempany CM, Kaus MR, Haker SJ, Wells WM III, Jolesz FA, Kikinis R (2004) Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 11:178–189
Lim JS (1990) Two-dimensional signal and image processing. Prentice Hall, Englewood Cliffs, p 548
Mendonca AM, Campilho A (2006) Segmentation of retinal blood vessels by combining the detection of centerlines and morphological reconstruction. IEEE Trans Med Imaging 25:1200–1213
Ma WY, Manjunath BS (2000) EdgeFlow: a technique for boundary detection and image segmentation. IEEE Trans Image Process 9:1375–1388
Johnson DH, Narayan S, Flask CA, Wilson DL (2010) Improved fat-water reconstruction algorithm with graphics hardware acceleration. J Magn Reson Imaging 31:457–465
Acknowledgments
The authors would like to acknowledge the expert technical help of Miss Catherine Doller and Dr. Scott Howell with tissue histology. This work was supported by the Case Center for Imaging Research. The research was also supported by grants Ohio Wright Center/BRTT, The Biomedical Structure, Functional and Molecular Imaging Enterprise (D.L.W), National Institutes of Health grants R42CA124270 (D.L.W.), T32EB007509 (K.E.S), R01-NS051520 (S.B.-K.), and R01-NS063971 (S.B.-K., D.L.W. and J.P.B.).
Conflict of Interest
Debashish Roy, David L. Wilson, Mohammed Q. Qutaish, and Kristin E. Sullivant are inventors of cryo-imaging technology licensed by CWRU. Debashish Roy and David L. Wilson are officers with ownership positions at BioInVision, Inc., which is commercializing cryo-imaging.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Whole brain visualization of blood vessels and tumor 1. 3D rendering of bright field volume of a 20-day-old tumor fades to show the results of the blood vessel enhancement algorithm (red) along with the results of the main tumor mass and dispersal cell detection algorithms (green and yellow, respectively). Rotation around the brain clearly shows the ability of the blood vessel enhancement algorithm to visualize blood vessels without the need for a contrast agent. (M4V 5814 kb)
LN-229 cell dispersal along blood vessels. ROI around LN-229 tumor at 20 days post-implantation shows blood vessels, main tumor mass, and dispersed cell clusters in red, green, and yellow, respectively. Dispersal along blood vessel is clearly shown by rotation and zooming (tumor 1). (M4V 5948 kb)
Rights and permissions
About this article
Cite this article
Qutaish, M.Q., Sullivant, K.E., Burden-Gulley, S.M. et al. Cryo-image Analysis of Tumor Cell Migration, Invasion, and Dispersal in a Mouse Xenograft Model of Human Glioblastoma Multiforme. Mol Imaging Biol 14, 572–583 (2012). https://doi.org/10.1007/s11307-011-0525-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-011-0525-z