Abstract
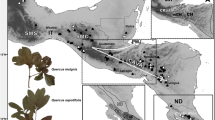
In widespread taxa in which hybridization is suspected of being an important aspect of species biology, the patterns and drivers of lineage diversification are not always clear. Here, we examine the patterns of species diversification in a monophyletic lineage of oaks endemic to western Mexico, a center of global oak diversity. This group of four species inhabits a variety of soil types and exhibits varying patterns of species distribution ranging from widespread to restricted, with the range bisected by the Trans-Mexican Volcanic Belt (TVB). Using chloroplast and nuclear microsatellites, we evaluated genetic diversity, diversification, and genetic group assignment across 49 populations. Chloroplast data identified 29 haplotypes in three distinct lineages, many of these found occurring on opposite sides of the TVB. Individual species were only loosely associated to specific chloroplast haplotypes but the distribution of shared haplotypes supports a one-time wider distribution. One lineage was highly divergent and geographically isolated, likely representing a case of chloroplast capture via hybridization and may indicate a range expansion of Racemiflorae into new territory. Nuclear gene diversity varied little across species and populations; however, differentiation and genetic assignment analysis was strongly structured geographically and tended to cluster with broad soil types. Soil specialist taxa produced homogeneous genetic structure while soil generalists showed varying patterns of mixed ancestry and high levels of admixture. Speciation appears driven by a combination of distance and edaphic factors leading to drift. Hybridization appears to be complementary, occurring most often in non-specific soil environments and in marginal areas, and contributes to a long-scale pattern of gene exchange as species ranges fluctuate with time.








Similar content being viewed by others
References
Aldrich PR, Michler CH, Sun W, Romero-Severson J (2002) Microsatellite markers for northern red oak (Fagaceae: Quercus rubra). Mol Ecol Notes 2:472–474
Arnold ML (1997) Natural hybridization and evolution. Oxford University Press, New York
Backs JR, Ashley MV (2016) Evolutionary history and gene flow of an endemic island oak: Quercus pacifica. Am J Bot 103:1–11
Bacon JR, Spellenberg R (1996) Hybridization in two distantly related Mexican black oaks Quercus conzattii and Quercus eduardii (Fagales: Quercus: section Lobatae). Sida 17:17–41
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Beatty GE, Montgomery WI, Spaans F, Tosh DG, Provan J (2016) Pure species in a continuum of genetic and morphological variation: sympatric oaks at the edge of their range. Ann Bot 117:541–549
Berrío JC, Hooghiemstra H, van Geel B, Ludlow-Wiechers B (2006) Environmental history of the dry forest biome of Guerrero, Mexico, and human impact during the last c. 2700 years. The Holocene 16:63–80
Burgarella C, Lorenzo Z, Jabbour-Zahab R, Lumaret R, Guichoux E, Petit RJ, Soto A, Gil L (2009) Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex). Heredity 102:442–452
Burger WC (1975) The species concept in Quercus. Taxon 24:45–50
Buschbom J, Yanbaev Y, Degen B (2011) Efficient long-distance gene flow into an isolated relict oak stand. J Hered 102:464–472
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Am J Hum Genet 19:233–257
Cavender-Bares J, Pahlich A (2009) Molecular, morphological, and ecological niche differentiation of sympatric sister oak species, Quercus virginiana and Q. geminata (Fagaceae). Am J Bot 96:1690–1702
Cavender-Bares J, González-Rodríguez A, Eaton DA, Hipp AA, Beulke A, Manos PS (2015) Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. Mol Ecol 24:3668–3687
Chávez-Álvarez MJ, Cerca M, Ferrari L (2012) Physical and geological description of the Nanchititla dyke swarm. Rev Mex Cienc Geol 29:551–571
Chybicki IJ, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100:106–113
Croizat L (1958) Panbiogeography. Caracas, published by the author
Croizat L, Nelson G, Rosen DE (1974) Centers of origin and related concepts. Syst Zool 23:265–267
Curtu AL, Gailing O, Finkeldey R (2007) Evidence for hybridization and introgression within a species-rich oak (Quercus spp.) community. BMC Evol Biol 7:218
Curtu AL, Gailing O, Finkeldey R (2009) Patterns of contemporary hybridization inferred from paternity analysis in a four-oak-species forest. BMC Evol Biol 9:284
Daghlian CP, Crepet WL (1983) Oak catkins, leaves and fruits from the Oligocene Catahoula Formation and their evolutionary significance. Am J Bot 70:639–649
Dumolin-Lapègue S, Kremer A, Petit RJ (1999) Are chloroplast and mitochondrial DNA variation species independent in oaks? Evolution 53:1406–1413
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics Online 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Ferrari L, Valencia-Moreno M, Bryan S (2007) Magmatism and tectonics of the Sierra Madre Occidental and its relation with the evolution of the western margin of North America. In: Alaniz-Álvarez SA, Nieto-Samaniego ÁF (eds) Geology of México: celebrating the centenary of the Geological Society of México: Geological Society of America special paper 422. Geological Society of America, Boulder, pp 1–39
Foll M, Gaggiotti OE (2006) Identifying the environmental factors that determine the genetic structure of populations. Genetics 174:875–891
Galicia L, Potvin C, Messierd C (2015) Maintaining the high diversity of pine and oak species in Mexican temperate forests: a new management approach combining functional zoning and ecosystem adaptability. Can J For Res 45:1358–1368
Gerber S, Chadœuf J, Gugerli F, Lascoux M, Buiteveld J, Cottrell J, Dounavi A, Fineschi S, Forrest LL, Fogelqvist J, Goicoechea PG, Svejgaard Jensen J, Salvini D, Vendramin GG, Kremer A (2014) High rates of gene flow by pollen and seed in oak populations across Europe. PLoS One 9:e85130
Gómez-Tuena A, Orozco-Esquivel MT, Ferrari L (2007) Igneous petrogenesis of the Trans-Mexican Volcanic Belt. In: Alaniz-Álvarez SA, Nieto-Samaniego ÁF (eds) Geology of México: celebrating the centenary of the Geological Society of México: Geological Society of America special paper 422. Geological Society of America, Boulder, pp 129–181
González-Rodríguez A, Arias DM, Valencia S, Oyama K (2004a) Morphological and RAPD analysis of hybridization between Quercus affinis and Q. laurina (Fagaceae), two Mexican red oaks. Am J Bot 91:401–409
González-Rodríguez A, Bain JF, Golden JL, Oyama K (2004b) Chloroplast DNA variation in the Quercus affinis-Quercus laurina complex in Mexico: geographical structure and associations with nuclear and morphological variation. Mol Ecol 13:3467–3476
Goudet J (1995) FSTAT (ver. 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet (1995)
Grivet D, Deguilloux M-F, Petit RJ, Sork VL (2006) Contrasting patterns of historical colonization in white oaks (Quercus spp.) in California and Europe. Mol Ecol 15:4085–4093
Hauser DA, Keuter A, McVay JD, Hipp AL, Manos PS (2017) The evolution and diversification of the red oaks of the California Floristic Province (Quercus section Lobatae, series Agrifoliae). Am J Bot 104:1581–1595
Herrera Arroyo ML (2005) Analisis de la morfologia foliar de tres especies de encinos mexicanos Quercus conzattii, Q. urbanii y Q. eduardii (Fagaceae: Lobatae), variacion, diferenciacion e hibridacion. MS Thesis, Universidad Nacional Autónoma de México, Mexico
Herrera-Arroyo ML, Sork VL, González-Rodríguez A, Rocha-Ramírez V, Vega E, Oyama K (2013) Seed-mediated connectivity among fragmented populations of Quercus castanea (Fagaceae) in a Mexican landscape. Am J Bot 100:1663–1671
Hintze R (1995) Number Cruncher Statistical Software [NCSS]. Kaysville, UT
Hipp AL (2015) Should hybridization make us skeptical of the oak phylogeny? Int Oaks 26:9–17
Hipp AL, Eaton DAR, Cavender-Bares J, Fitzek E, Nipper R, Manos PS (2014) A framework phylogeny of the American Oak clade based on sequenced RAD data. PLoS One 9:e93975
Hipp AL, Manos PS, González-Rodríguez HM, Kaproth M, McVay JD, Valencia Avalos S, Cavender-Bares J (2017) Sympatric parallel diversification of major oak clades in the Americas and the origins of Mexican species diversity. New Phytol 217:439–452
Hooghiemstra H (2006) Immigration of oak into Northern South America: a paleo-ecological document. In: Kapelle M (ed) Ecology and conservation of neotropical montane oak forests, ecological studies, vol 185. Springer-Verlag, Berlin, pp 17–28
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:801–1806
Jiménez P, López de Heredia U, Collada C, Lorenzo Z, Gil L (2004) High variability of chloroplast DNA in three Mediterranean evergreen oaks indicates complex evolutionary history. Heredity 93:510–515
Jørgensen S, Mauricio R (2005) Hybridization as a source of evolutionary novelty: leaf shape in Hawaiian composite. Genetica 123:171–179
Kremer A, Kleinschmit J, Cottrell J, Cundall EP, Deans JD, Ducousso A, König AO, Lowe AJ, Munro RC, Petit RJ, Stephan BR (2002) Is there a correlation between chloroplastic and nuclear divergence, or what are the roles of history and selection on genetic diversity in European oaks? For Ecol Manag 156:75–87
Langella O (1999) POPULATIONS 1.2.31: population genetic software, individuals or populations distances, phylogenetic trees. Available from http://bioinformatics.org/populations/
Lefort F, Douglas GC (1999) An efficient micro-method of DNA isolation from mature leaves of four hardwood tree species Acer, Fraxinus, Prunus, and Quercus. Ann For Sci 56:59–263
Lepais O, Petit RJ, Guichoux E, Lavabre JE, Alberto F, Kremer A, Gerber S (2009) Species relative abundance and direction of introgression in oaks. Mol Ecol 18:2228–2242
Lexer C, Kremer A, Petit RJ (2006) Shared alleles in sympatric oaks: recurrent gene flow is a more parsimonious explanation than ancestral polymorphism. Mol Ecol 15:2007–2012
Lind-Riehl JF, Sullivan AR, Gailing O (2014) Evidence for selection on a CONSTANS-like gene between two red oak species. Ann Bot 113:967–975
Magri D, Fineschi S, Bellarosa R, Buonamici A, Sebastiani F, Schirone B, Simeone MC, Vendramin GG (2007) The distribution of Quercus suber chloroplast haplotypes matches the palaeogeographical history of the western Mediterranean. Mol Ecol 16:5259–5266
Manos PS, Doyle JJ, Nixon KC (1999) Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae). Mol Phylogenet Evol 12:333–349
Martinez M (1966) Los encinos de México. XIV. Anales del Instituto de Biología de la Universidad Nacional de México 37:81–95
Martínez-Hernández E, Ramírez-Arriaga E (2006) Tertiary palynofloristic correlations between Mexican formations with emphasis in dating the Balsas Group. In: Vega FJ, Nyborg TG, Perrilliat MC, Montellano-Ballesteros M, Cevallos-Ferriz SRS, Quiroz-Barroso SA (eds) Studies on Mexican paleontology. Springer, Dordrecht, pp 19–45
McVaugh R (1974) Fagaceae. Flora Novo-Galiciana. Contrib Univ Mich Herb 12 (pt.1, no. 3):1–93
McVay JD, Hipp AL, Manos PS (2017) A genetic legacy of introgression confounds phylogeny and biogeography in oaks. Proc R Soc B 284:20170300
Morán-Zenteno DJ, Cerca M, Keppie JD (2007) The Cenozoic tectonic and magmatic evolution of southwestern Mexico: advances and problems of interpretation. In: Alaniz-Álvarez SA, Nieto-Samaniego ÁF (eds) Geology of México: celebrating the centenary of the Geological Society of México: Geological Society of America special paper 422. Geological Society of America, Boulder, pp 71–91
Morrone JJ, Crisci JV (1995) Historical biogeography: introduction to methods. Annu Rev Ecol Syst 26:373–401
Muir G, Schlötterer C (2005) Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp.). Mol Ecol 14:549–561
Muir G, Fleming CC, Schlötterer C (2000) Species status of hybridizing oaks. Nature 405:1016
Muller CH (1952) Ecological control of hybridization in Quercus: a factor in the mechanism of evolution. Evolution 6:147–161
Nixon KC (1993) The genus Quercus in Mexico. In: Ramammoorthy TP, Bye R, Lot A, Fa J (eds) Biological diversity of Mexico: origins and distribution. Oxford University Press, New York, pp 447–458
Ortego J, Noguerales V, Gugger PF, Sork VL (2015) Evolutionary and demographic history of the Californian scrub white oak species complex: an integrative approach. Mol Ecol 24:6188–6208
Ortego J, Gugger PF, Sork VL (2017) Impacts of human-induced environmental disturbances on hybridization between two ecologically differentiated Californian oak species. New Phytol 213:942–955
Owusu SA, Sullivan AR, Weber JA, Hipp AL, Gailing O (2015) Taxonomic relationships and gene flow in four North American Quercus species (Quercus section Lobatae). Syst Bot 40:510–521
Palmer EJ (1948) Hybrid oaks of North America. J Arnold Arbor 29:1–48
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Peñaloza-Ramírez JM, González-Rodríguez A, Mendoza-Cuenca L, Caron H, Kremer A, Oyama K (2010) Interspecific gene flow in a multispecies oak hybrid zone in the Sierra Tarahumara of Mexico. Ann Bot 105:389–399
Petit RJ, Csaikl UM, Bordács S, Burg K, Coart E, Cottrell J, van Dam B, Deans JD, Dumolin-Lapègue S, Fineschi S, Finkeldey R, Gillies A, Glaz I, Goicoechea PG, Jensen JS, König AO, Lowe AJ, Madsen SF, Mátyás G, Munro RC, Olalde M, Pemonge MH, Popescu F, Slade D, Tabbener H, Taurchini D, de Vries SGM, Ziegenhagen B, Kremer A (2002) Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. For Ecol Manag 156:5–26
Petit RJ, Bodenes C, Ducousso A, Roussel G, Kremer A (2003) Hybridization as a mechanism of invasion in oaks. New Phytol 161:151–164
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Pritchard JK, Stephens P, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ramos-Ortiz S, Oyama K, Rodríguez-Correa H, González-Rodríguez A (2015) Geographic structure of genetic and phenotypic variation in the hybrid zone between Quercus affinis and Q. laurina in Mexico. Plant Species Biol 31:219–232
Rattenbury JA (1962) Cyclic hybridization as a survival mechanism in the New Zealand forest flora. Evolution 16:348–363
Rodríguez-Correa H, Oyama K, MacGregor-Fors I, González-Rodríguez A (2015) How are oaks distributed in the neotropics? A perspective from species turnover, areas of endemism, and climatic niches. Int J Plant Sci 176:222–231
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rubio de Casas R, Cano E, Balaguer L, Pérez-Corona E, Manrique E, García-Verdugo C, Vargas P (2007) Taxonomic identity of Quercus coccifera L. in the Iberian Peninsula is maintained in spite of widespread hybridization, as revealed by morphological, ISSR and ITS sequence data. Flora 202:488–499
Ruiz-Sánchez E, Rodríguez-Gómez F, Sosa V (2012) Refugia and geographic barriers of populations of the desert poppy, Hunnemannia fumariifolia (Papaveraceae). Org Divers Evol 26:991–1010
Rzedowski J (1978) Vegetación de México. Limusa, México
Scareli-Santos C, Herrera-Arroyo ML, Sánchez-Mondragón ML, González-Rodríguez A, Bacon J, Oyama K (2007) Comparative analysis of micromorphological characters in two distantly related Mexican oaks, Quercus conzattii and Q. eduardii (Fagaceae), and their hybrids. Brittonia 59:37–48
Schlötterer C (2002) A microsatellite-based multilocus screen for the identification of local selective sweeps. Genetics 160:753–763
Sebastiani F, Carnevale C, Vendramin GG (2004) A set of mono- and dinucleotide chloroplast microsatellites in Fagaceae. Mol Ecol Notes 4:259–261
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82:561–573
Sork VL, Stowe KA, Hochwender C (1993) Evidence for local adaptation in closely adjacent subpopulations of northern red oak (Quercus rubra L.) expressed as resistance to leaf herbivores. Am Nat 142:928–936
Sosa V, Ruiz-Sanchez E, Rodriguez-Gomez FC (2009) Hidden phylogeographic complexity in the Sierra Madre Oriental: the case of the Mexican tulip poppy Hunnemannia fumariifolia (Papaveraceae). J Biogeogr 36:18–27
Spellenberg R, Bacon JR (1996) Taxonomy and distribution of a natural group of black oaks of Mexico (Quercus, section Lobatae, subsection Racemiflorae). Syst Bot 21:85–99
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Torres-Miranda A, Luna-Vega I, Oyama K (2011) Conservation biogeography of red oaks (Quercus section Lobatae) in Mexico and Central America. Am J Bot 98:290–305
Tovar-Sánchez E, Oyama K (2004) Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: morphological and molecular evidence. Am J Bot 91:1352–1363
Trelease W (1921) A natural group of unusual black oaks. Proc Am Philos Soc 60:31–33 + 3 plates
Vähä JP, Primmer CR (2006) Efficiency of model based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol 15:63–72
Valbuena-Carabaña M, González-Martínez SC, Sork VL, Collada C, Soto A, Goicoechea PG, Gil L (2005) Gene flow and hybridisation in a mixed oak forest (Quercus pirenaica Willd. and Quercus petraea (Matts.) Liebl.) in central Spain. Heredity 95:457–465
Valencia-A S (2004) Diversidad del género Quercus (Fagaceae) en México. Bol Soc Bot Méx 75:33–53
Valencia-Cuevas L, Piñero D, Mussali-Galante P, Valencia-Ávalos S, Tovar-Sánchez E (2014) Effect of a red oak species gradient on genetic structure and diversity of Quercus castanea (Fagaceae) in Mexico. Tree Genet Genomes 10:641–652
Valencia-Cuevas L, Mussali-Galante P, Piñero D, Castillo-Mendoza E, Rangel-Altamirano G, Tovar-Sánchez E (2015) Hybridization of Quercus castanea (Fagaceae) across a red oak species gradient in Mexico. Plant Syst Evol 301:1085–1097
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHEKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Van Valen L (1976) Ecological species, multispecies, and oaks. Taxon 25:233–239
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whittemore AT, Schaal BA (1991) Interspecific gene flow in sympatric oaks. Proc Natl Acad Sci U S A 88:2540–2544
Zeng Y-F, Liao W-J, Petit RJ, Zhang D-Y (2010) Exploring species limits in two closely related Chinese oaks. PLoS One 5:e15529
Acknowledgments
The authors thank María Luisa Herrera Arroyo for providing selected population samples of Racemiflorae, Juan Peñaloza-Ramírez and Enrique Pascual-Alvarado for assistance in field collections, and Rennan Moreira for assistance in DNA extraction. We would also like to thank Andrew Hipp and three anonymous reviewers for their useful suggestions on the manuscript.
Funding
This work was supported by the Consejo Nacional de Ciencia y Tecnología and the Secretaría de Medio Ambiente y Recursos Naturales - Mexico (CONACYT-SEMARNAT) [grant 2004-C01–97 to K.O.], the Dirección General de Asuntos del Personal Académico of the Universidad Nacional Autónoma de México and the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (DGAPA-PAPIIT, UNAM) [grant IN229803 to K.O.], and DGAPA, UNAM Postdoctoral Fellowships to R.A.M and A.C.C-P.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data archiving statement
All microsatellite data upon which this study is based is archived at the Dryad Digital Repository. https://doi.org/10.5061/dryad.8dq043h.
Additional information
Communicated by J. Wright
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
a Mean L(K) ± SD (=Ln P(D)) for 5 replicate runs at each of 10 proposed clusters (K). b Rate of change of the likelihood distribution (L′(K) ± SD) for 10 proposed clusters (K). c Absolute values of the second order rate of change of the likelihood distribution (|L′′(K)| ± SD). d ΔK. Most supported number of groups indicated by tallest peak (K = 8) (JPG 101 kb)
Rights and permissions
About this article
Cite this article
McCauley, R.A., Cortés-Palomec, A.C. & Oyama, K. Species diversification in a lineage of Mexican red oak (Quercus section Lobatae subsection Racemiflorae)—the interplay between distance, habitat, and hybridization. Tree Genetics & Genomes 15, 27 (2019). https://doi.org/10.1007/s11295-019-1333-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-019-1333-x