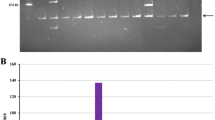
Abstract
Tea is a popular and natural non-alcoholic beverage, and is produced from fresh leaves of Camellia sinensis. Tea leaves contain many bioactive compounds that have significant health benefits. We constructed a high quality bacterial artificial chromosome (BAC) library by using the fresh petals of C. sinensis “Shuchazao” for genome sequencing and improvement of genomic assembly. BAC library is still a significant tool for studies of functional genomes and preservation of precious genetic resources. The BAC library contains 161,280 clones with an average insert size of 113 kb, which represents approximately 6.2-fold coverage of haploid genome equivalents of C. sinensis. We characterized 20 complete BAC clones and 738 BAC end sequences (BESs) ranging from 105 to 917 bp. In addition, we predicted cis-regulatory elements of LAR (leucoanthocyanidin reductase), TCS (caffeine synthase), and TS (theanine synthetase) involved in tea characteristic metabolite synthesis and identified a larger number of light-responsive cis-acting elements in these three genes. Meanwhile, we analyzed alternative splicing of these three genes. Furthermore, 12 pairs of SSR primers were successfully amplified in tea plant DNA. The tea BAC library was a critical resource to accomplish de novo whole-genome sequencing, accelerate gene discovery and enhance molecular breeding of C. sinensis.









Similar content being viewed by others
References
Allouis S, Qi X, Lindup S, Gale M, Devos K (2001) Construction of a BAC library of pearl millet, Pennisetum glaucum. Theor Appl Genet 102:1200–1205
Ammiraju JS, Meizhong L, Goicoechea JL, Wenming W, Dave K, Christopher M et al (2006) The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res 16:140–147
Ammiraju JS, Lu F, Abhijit S, Yeisoo Y, Song X, Jiang N et al (2008) Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell 20:3191–3209
Anistoroaei R, ten Hallers B, Nefedov M, Christensen K, de Jong P (2011) Construction of an American mink Bacterial Artificial Chromosome (BAC) library and sequencing candidate genes important for the fur industry. BMC Genomics 12:354
Auvichayapat P, Montira P, Oratai T, Narong A (2008) Effectiveness of green tea on weight reduction in obese Thais: a randomized, controlled trial. Physiol Behav 93:486–491
Cao W, Fu B, Wu K, Li N, Zhou Y, Gao Z et al (2014) Construction and characterization of three wheat bacterial artificial chromosome libraries. Int J Mol Sci 15:21896–21912
Cenci A, Chantret N, Kong X, Gu Y, Andersonet OD, Fahima T, Distelfeld A, Dubcovsky J (2003) Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp. durum). Theor Appl Genet 107:931–939
Chen, P. (2010). Shading effects and influences on photosynthesis and quality component of tea in tea plantation. Hunan Agricultural University
Chen M, SanMiguel P, De Oliveira A, Woo S-S, Zhang H, Wing R, Bennetzen J (1997) Microcolinearity in sh2-homologous regions of the maize, rice, and sorghum genomes. Proc Natl Acad Sci 94:3431–3435
Chen M, Gernot P, Barbazuk B, Goicoechea J, Barbara B, Fang G et al (2002) An integrated physical and genetic map of the rice genome. Plant Cell 14:537–545
David P, Sévignac M, Thareau V, Catillon Y, Kami J, Gepts P, Thierry L, Vale’rie G (2008) BAC end sequences corresponding to the B4 resistance gene cluster in common bean: a resource for markers and synteny analyses. Mol Gen Genomics 280:521–533
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phredII Error probabilities. Genome Res 8:186–194
Feng C, Bluhm BH, Correll JC (2015) Construction of a spinach bacterial artificial chromosome (BAC) library as a resource for gene identification and marker development. Plant Molecular. Biology Reporter:1–10
Frary A, Hamilton CM (2001) Efficiency and stability of high molecular weight DNA transformation: an analysis in tomato. Transgenic Res 10:121–132
Gordon D (2003) Viewing and editing assembled sequences using Consed. Current protocols in bioinformatics 11.12(11-11.12):43
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5:225–225
Hodgson JM, Croft KD, Woodman RJ, Puddey IB, Fuchs D, Draijer R, Lukoshkova E, Head GA (2013) Black tea lowers the rate of blood pressure variation: a randomized controlled trial. Am J Clin Nutr 97:943–950
Hollman PC, Feskens EJ, Katan MB (1999) Tea flavonols in cardiovascular disease and cancer epidemiology. Proc Soc Exp Biol Med 220:198–202
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Huang H, Tong Y, Zhang Q-J, Gao L-Z (2013) Genome size variation among and within camellia species by using flow cytometric analysis. PLoS One 8:e64981
Initiative AG (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796
Janda J, Šafář J, Kubaláková M, Jet B, Kovářová P, Suchánková P et al (2006) Advanced resources for plant genomics: a BAC library specific for the short arm of wheat chromosome 1B. Plant J 47:977–986
Jia X, Deng Y, Sun X, Liang L, Ye X (2015) Characterization of the global transcriptome using Illumina sequencing and novel microsatellite marker information in seashore paspalum. Genes Genomics 37:77–86
Jin Q, Chen Z, Sun W, Lin F, Xue Z, Huang Y, Tang X (2016) Cloning and Bioinformatical analysis of anthocyanin synthase gene and its promoter in Camellia sinensis. J Tea Sci 36:219–228
Kofler R, Schlötterer C, Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685
Kuehnbaum NL, Kormendi A, Britz-McKibbin P (2013) Multisegment injection-capillary electrophoresis-mass spectrometry: a high-throughput platform for metabolomics with high data fidelity. Anal Chem 85:10664–10669
Li P (2005) International Rice Genome Sequencing Project. 2005. The map-based sequence of the rice genome. Nature 436:793–800
Liang Y, Ma W, Lu J, Wu Y (2001) Comparison of chemical compositions of Ilex latifolia Thumb and Camellia sinensis L. Food Chem 75:339–343
Lin J, Kudrna D, Wing RA (2011) Construction, characterization, and preliminary BAC-end sequence analysis of a bacterial artificial chromosome library of the tea plant (Camellia sinensis) J Biomed Biotechnol 2011:476723
Lin H, Xia P, Wing RA, Zhang Q, Luo M (2012) Dynamic intra-japonica subspecies variation and resource application. Mol Plant 5:218–230
Liu Y, Gao L, Liu L, Yang Q, Lu Z, Nie Z, Wang Y, Xia T (2012) Purification and characterization of a novel galloyltransferase involved in catechin galloylation in the tea plant (Camellia sinensis). J Biol Chem 287:44406–44417
Liu J, Yuan D, Si H, Pang X, Tang X, Yang J (2013a) Effects of shading on ingredients of tea shoots in different seasons. Southwest China J Agric Sci 26:115–118
Liu C, Guo Y, Lu T, Wu H, Na R, Li X, Guan W, Ma Y (2013b) Construction and preliminary characterization analysis of Wuzhishan miniature pig bacterial artificial chromosome library with approximately 8-fold genome equivalent coverage. Biomed Res Int
Lu F, Ammiraju JS, Sanyal A, Zhang S, Song R, Chen J, Li G, Sui Y, Song X et al (2009) Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes. Proc Natl Acad Sci 106:2071–2076
Luo M, Wing RA (2003) An improved method for plant BAC library construction. In: Plant functional genomics. Springer, pp 3–19
Ma J, Zhou Y, Ma C, Yao M, Jin J, Wang X, Chen L (2010) Identification and characterization of 74 novel polymorphic EST-SSR markers in the tea plant, Camellia sinensis (Theaceae). Am J Bot 97:e153–e156
Mahmood T, Akhtar N, Khan BA (2010) The morphology, characteristics, and medicinal properties of Camellia sinensis’ tea. J Med Plants Res 4:2028–2033
Mamati GE, Liang Y, Lu J (2006) Expression of basic genes involved in tea polyphenol synthesis in relation to accumulation of catechins and total tea polyphenols. J Sci Food Agric 86:459–464
Matsuura T, Kakuda T (1990) Effects of precursor, temperature, and illumination on Theanine accumulation in tea callus. Agric Biol Chem 37:2033–2051
Messing J, Llaca V (1998) Importance of anchor genomes for any plant genome project. Proc Natl Acad Sci 95:2017–2020
Nilmalgoda SD, Cloutier S, Walichnowski AZ (2003) Construction and characterization of a bacterial artificial chromosome (BAC) library of hexaploid wheat (Triticum aestivum L.) and validation of genome coverage using locus-specific primers. Genome 46:870–878
O’Sullivan DM, Ripoll P, Rodgers M, Edwards K (2001) A maize bacterial artificial chromosome (BAC) library from the European flint inbred line F2. Theor Appl Genet 103:425–432
Pan Y, Deng Y, Lin H, Kudrna DA, Wing RA, Li L, Zhang Q, Luo M (2014) Comparative BAC-based physical mapping of Oryza sativa ssp. indica var. 93–11 and evaluation of the two rice reference sequence assemblies. Plant J 77:795-805
Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. Bioinforma Methods Protocol:365–386
Sanmiguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot 82:37–44
Sasazuki S, Tamakoshi A, Matsuo K, Ito H, Wakai K, Nagata C, Mizoue T, Tanaka K, Tsuji I, Inoue M, Tsugane S (2012) Green tea consumption and gastric cancer risk: an evaluation based on a systematic review of epidemiologic evidence among the Japanese population. Jpn J Clin Oncol 42:335–346
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Schulte D, Ariyadasa R, Shi B, Fleury D, Saski C, Atkins M, Pieter D, Wu C, Andreas G, Peter L, Nils S (2011) BAC library resources for map-based cloning and physical map construction in barley (Hordeum vulgare L.) Bmc Genomics 12:247
Shi C, Yang H, Wei C, Yu O, Zhang JC, Sun J, Li Y, Chen Q, Xia T, Wan X (2011a) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12:131
Shi X, Zeng H, Xue Y, Luo M (2011b) A pair of new BAC and BIBAC vectors that facilitate BAC/BIBAC library construction and intact large genomic DNA insert exchange. Plant Methods 7:33
Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M (1992) Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci 89:8794–8797
Song X, Goicoechea J, Ammiraju J, Luo M, He R, Lin J et al (2011) The 19 genomes of Drosophila: a BAC library resource for genus-wide and genome-scale comparative evolutionary research. Genetics 187:1023–1030
Taniguchi F, Hirai, N. and Yamaguchi, S, (2006) Estimation of the genome size of tea (Camellia sinensis), camellia (C. japonica), and their interspecific hybrids by flow cytometry. Journal of the Remote Sensing Society of Japan (Japan):1-7
Tomkins J, Davis G, Main D, Yim Y, Duru N, Musket T, Goicoechea J, Frisch D, Coe E, Wing R (2002) Construction and characterization of a deep-coverage bacterial artificial chromosome library for maize. Crop Sci 42:928–933
Trapnell C, Pachter L, Salzberg S (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Wang Q, Dooner HK (2012) Dynamic evolution of bz orthologous regions in the Andropogoneae and other grasses. Plant J 72:212–221
Wang GL, Holsten TE, Song WY, Wang HP, Ronald PC (1995) Construction of a rice bacterial artificial chromosome library and identification of clones linked to the Xa-21 disease resistance locus. Plant J 7:525–533
Wang Y, Gao L, Shan Y, Liu Y, Tian Y, Xia T (2012) Influence of shade on flavonoid biosynthesis in tea ( Camellia sinensis (L.) O. Kuntze). Sci Hortic 141:7–16
Wang C, Shi X, Liu L, Li H, Ammiraju J, Kudrna D, Xiong W, Wang H et al (2013a) Genomic resources for gene discovery, functional genome annotation, and evolutionary studies of maize and its close relatives. Genetics 195:723–737
Wang X, Zhao Q, Ma C, Zhang Z, Cao H, Kong Y (2013b) Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 14:415
Wang X, Liu Q, Wang H, Luo C-X, Wang G, Luo M (2013c) A BAC based physical map and genome survey of the rice false smut fungus Villosiclava virens. BMC Genomics 14:883
Wang X, Kudrna D, Pan Y, Wang H, Liu L, Lin H, Zhang J, Song X et al (2014a) Global genomic diversity of Oryza sativa varieties revealed by comparative physical mapping. Genetics 196:937–949
Wang Y, Xu Y, Gao L, Yu O, Wang X, He X, Jiang X, Liu Y, Xia T (2014b) Functional analysis of Flavonoid 3′ 5′-hydroxylase from ea plant (Camellia sinensis): critical role in the accumulation of catechins. BMC Plant Biol 14:347
Wei F, Zhang J, Zhou S, He R, Schaeffer M, Collura K et al (2009) The physical and genetic framework of the maize B73 genome. PLoS Genet 5:e1000715
Woo S-S, Jiang J, Gill BS, Paterson AH, Wing RA (1994) Construction and characterization of bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res 22:4922–4931
Wu C, Nimmakayala P, Santos F, Springman R, Scheuring C, Meksem K, Lightfoot D, Zhang H (2004) Construction and characterization of a soybean bacterial artificial chromosome library and use of multiple complementary libraries for genome physical mapping. Theor Appl Genet 109:1041–1050
Wu H, Chen D, Li J, Yu B, Qiao X, Huang H, He Y (2013) De novo characterization of leaf transcriptome using 454 sequencing and development of EST-SSR markers in tea (Camellia sinensis). Plant Mol Biol Report 31:524–538
Wu Z, Li X, Liu Z, Xu Z, Zhuang J (2014) De novo assembly and transcriptome characterization: novel insights into catechins biosynthesis in Camellia sinensis. BMC Plant Biol 14:277
Xia Z, Wu H, Watanabe S, Harada K (2014) Construction and targeted retrieval of specific clone from a non-gridded soybean bacterial artificial chromosome library. Anal Biochem 444:38–40
Xia E, Zhang H, Sheng J, Li K, Zhang Q, Kim C, et al (2017) The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Molecular Plant
Yen G, Chen H (1995) Antioxidant activity of various tea extracts in relation to their antimutagenicity. J Agric Food Chem 43:27–32
Yim Y, Davis G, Duru N, Musket T, Linton E, Messing J, McMullen M, Soderlund C, Polacco M, Gardiner J, Coe E (2002) Characterization of three maize bacterial artificial chromosome libraries toward anchoring of the physical map to the genetic map using high-density bacterial artificial chromosome filter hybridization. Plant Physiol 130:1686–1696
Yu J, Wang J, Lin W, Li S, Li H, Zhou J et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3:e38
Zhang H, Xia E, Huang H, Jiang J, Liu B, Gao L (2015) De novo transcriptome assembly of the wild relative of tea tree (Camellia taliensis) and comparative analysis with tea transcriptome identified putative genes associated with tea quality and stress response. BMC Genomics 16:298
Zhu H, Choi S, Johnston AK, Wing RA, Dean RA (1997) A large-insert (130 kbp) bacterial artificial chromosome library of the rice blast fungus Magnaporthe grisea: genome analysis, contig assembly, and gene cloning. Fungal Genet Biol 21:337–347
Acknowledgments
This work received financial support from the Science and Technology Project of AnHui Province, China (Project 13Z03012), Tea Genome Project of AnHui Province, China, the Special Innovative Province Construction in Anhui province in 2015 (15czs08032), the Central Guiding the Science and Technology Development of the Local (2016080503B024), the Major Project of Chinese National Programs for Fundamental Research and Development (2012CB722903), the Natural Science Foundation of Anhui Province (No.1608085QC60), and the Youth Foundation of Anhui Agricultural University (2016ZR012). We appreciate Chun Liu (Beijing Genome Institute at Shenzhen, China) for technical support and analysis. We are grateful to the unknown editor at the elixigen editing service (ID151023-6181) for the English polishing.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiment: WC and SJ; analyzing data: WL, WH, TL, JY, LZ, YH, and DW; experiment: SL, DZ, SY, HB, WQ, and LM. TL and WH contributed to writing the text.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Communicated by W.-W. Guo
Data achieving statement
The raw data of tea plant BAC are now are available in the NCBI SRA under project accession number PRJNA385558. The experiment accession numbers were SRX2982355. The BAC sequences of tea plant will be submitted to the European Nucleotide Archive database (ENA; http://www.ebi.ac.uk/ena) under project number accession PRJEB21650 if the manuscript is accepted for publication in the tree genetics and genomes prior to publication.
The Illumina RNA-seq data used in this study has been deposited in the NCBI SRA (http://trace.ncbi.nlm.nih.gov/Traces/sra) with accessions SRR1928149.
Electronic supplementary material
Supplementary Figures S1
(DOCX 72 kb)
Supplementary Figures S2
(DOCX 498 kb)
Supplementary Figures S3
(DOCX 93 kb)
Supplementary Figures S4
(DOCX 75 kb)
ESM 1
(XLSX 180 kb)
Rights and permissions
About this article
Cite this article
Tai, Y., Wang, H., Wei, C. et al. Construction and characterization of a bacterial artificial chromosome library for Camellia sinensis . Tree Genetics & Genomes 13, 89 (2017). https://doi.org/10.1007/s11295-017-1173-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1173-5