Abstract
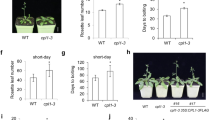
The transition to flowering is a major developmental switch in flowering plants. The nuclear RNA-binding protein FCA responds to seasonal signals and abscisic acid (ABA), which can control the flowering time via ambient temperature and autonomous pathways. Citrus FCA ortholog (PtFCA) has been isolated and characterized from precocious trifoliate orange (Poncirus trifoliata L. Raf). Three alternatively spliced transcripts of PtFCA (PtFCA1, PtFCA2, and PtFCA3) were isolated. The expression pattern of PtFCA indicated that it may be involved in phase transition in precocious trifoliate orange. A functional complementation experiment of PtFCA indicated that PtFCA1 partially rescued the late-flowering phenotype of the fca-1 mutant. There was no influence on flowering time of transgenic Arabidopsis by PtFCA3 as compared with PtFCA2, which exhibits delayed flowering time in a fca-1 background. Meanwhile, these three transcripts also showed different abilities to regulate root development in the fca-1 background. The study of protein–protein interactions suggested that PtFCA may form higher order complexes with PtFY and PtNF-YA7 to regulate timing of the transition from the vegetative to reproductive phase in precocious trifoliate orange. ABA and ambient temperature treatments changed the expression of PtFCA and interaction protein. These findings reveal that PtFCA may play important roles in flowering time and root development of precocious trifoliate orange through the formation of multiple protein complexes.






Similar content being viewed by others
References
Abou-Elwafa SF, Buttner B, Chia T, Schulze-Buxloh G, Hohmann U, Mutasa-Gottgens E, Jung C, Muller AE (2011) Conservation and divergence of autonomous pathway genes in the flowering regulatory network of Beta vulgaris. J Exp Bot 62:3359–3374
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14:S111–S130
Amasino RM, Michaels SD (2010) The timing of flowering. Plant Physiol 154:516–520
Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33:168–171
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Davenport T (1990) Citrus flowering. Hortic Rev 12:349–408
Gao YG, Yang H, Zhao J, Jiang YJ, Hu HY (2014) Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin. Structure 22:378–386
Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, Taylor L, Yeung B, Vassilovski G, Amin M, Chen F, Matskova L, Winberg G, Ernberg I, Linding R, O’Donnell P, Starostine A, Keller W, Metalnikov P, Stark C, Pawson T (2005) WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol 25:7092–7106
Jaeger KE, Graf A, Wigge PA (2006) The control of flowering in time and space. J Exp Bot 57:3415–3418
Jang YH, Lee JH, Park H-Y, Kim S-K, Lee B-Y, Suh MC, Kim J-K (2009a) OsFCA transcripts show more complex alternative processing patterns than its Arabidopsis counterparts. J Plant Biol 52:161–166
Jang YH, Park HY, Kim SK, Lee JH, Suh MC, Chung YS, Paek KH, Kim JK (2009b) Survey of rice proteins interacting with OsFCA and OsFY proteins which are homologous to the Arabidopsis flowering time proteins, FCA and FY. Plant Cell Physiol 50:1479–1492
Risk JM, Macknight RC, Da CL (2008) FCA does not bind abscisic acid. Nature 456:E5–E6
Jung JH, Seo PJ, Ahn JH, Park CM (2012) Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J Biol Chem 287:16007–16016
Khan MR, Ai XY, Zhang JZ (2014) Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip Rev RNA 5:347–359
Kriechbaumer V, Wang P, Hawes C, Abell BM (2012) Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 70:292–302
Kumar S, Jiang S, Jami SK, Hill RD (2011) Cloning and characterization of barley caryopsis FCA. Physiol Plant 143:93–106
Lee JH, Cho YS, Yoon HS, Suh MC, Moon J, Lee I, Weigel D, Yun CH, Kim JK (2005) Conservation and divergence of FCA function between Arabidopsis and rice. Plant Mol Biol 58:823–838
Lee HJ, Jung JH, Llorca LC, Kim SG, Lee S, Baldwin IT, Park CM (2014) FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat Commun 5:5473–5473
Li ZM, Zhang JZ, Mei L, Deng XX, Hu CG, Yao JL (2010) PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Mol Biol 74:129–142
Liang S, Zhu W, Xiang W (1999) Precocious trifoliate orange (Poncirus trifoliata (L.) Raf.) biology characteristic and its stock experiment. ZheJiang Citrus 16:2–4
Lindqvist C, Laakkonen L, Albert VA (2007) Polyglutamine variation in a flowering time protein correlates with island age in a Hawaiian plant radiation. BMC Evol Biol 7:105
Liu D, Cai X (2013) OsRRMh, a spen-like gene, plays an important role during the vegetative to reproductive transition in rice. J Integr Plant Biol 55:876–887
Liu F, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28:398–407
Macknight R (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89:737–745
Macknight R (2002) Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14:877–888
Nishikawa F, Endo T, Shimada T, Fujii H, Shimizu T, Omura M, Ikoma Y (2007) Increased CiFT abundance in the stem correlates with floral induction by low temperature in Satsuma mandarin (Citrus unshiu Marc.). J Exp Bot 58:3915–3927
Papadopoulos C, Arato K, Lilienthal E, Zerweck J, Schutkowski M, Chatain N, Muller-Newen G, Becker W, de la Luna S (2011) Splice variants of the dual specificity tyrosine phosphorylation-regulated kinase 4 (DYRK4) differ in their subcellular localization and catalytic activity. J Biol Chem 286:5494–5505
Quesada V (2003) Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time.pdf. EMBO J 22:3142–3152
Razem FA, El-Kereamy A, Abrams SR, Hill RD (2006) The RNA-binding protein FCA is an abscisic acid receptor. Nature 439:290–294
Remy E, Cabrito TR, Baster P, Batista RA, Teixeira MC, Friml J, Sa-Correia I, Duque P (2013) A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25:901–926
Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A (2007) A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19:2370–2390
Sarnowski TJ, Swiezewski S, Pawlikowska K, Kaczanowski S, Jerzmanowski A (2002) AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res 30:3412–3421
Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF 3rd (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149:625–641
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113:777–787
Simpson GG, Laurie RE, Dijkwel PP, Quesada V, Stockwell PA, Dean C, Macknight RC (2010) Noncanonical translation initiation of the Arabidopsis flowering time and alternative polyadenylation regulator FCA. Plant Cell 22:3764–3777
Siriwardana CL, Kumimoto RW, Jones DS, Holt BF (2014) Gene family analysis of the Arabidopsis NF-YA transcription factors reveals opposing abscisic acid responses during seed germination. Plant Mol Biol Report 32:971–986
Sorin C, Declerck M, Christ A, Blein T, Ma L, Lelandais-Briere C, Fransiska Njo M, Beeckman T, Crespi M, Hartmann C (2014) A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol 202:1197–1211
Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68:2013–2037
Sun F, Liu C, Zhang C, Qi W, Zhang X, Wu Z, Kong D, Wang Q, Shang H, Qian X, Li F, Yang J (2011) A conserved RNA recognition motif (RRM) domain of Brassica napus FCA improves cotton fiber quality and yield by regulating cell size. Mol Breed 30:93–101
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tan FC, Swain SM (2007) Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol Plant 131:481–495
von Arnim A (2007) Subcellular localization of GUS- and GFP-tagged proteins in onion epidermal cells. Cold Spring Harb Protoc 2007: pdb. prot4689
Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438
Zhang J-Z, Ai X-Y, Sun L-M, Zhang D-L, Guo W-W, Deng X-X, Hu C-G (2011a) Molecular cloning and functional characterization of genes associated with flowering in citrus using an early-flowering trifoliate orange (Poncirus trifoliata (L.) Raf.) mutant. Plant Mol Biol 76:187–204
Zhang J-Z, Ai X-Y, Sun L-M, Zhang D-L, Guo W-W, Deng X-X, Hu C-G (2011b) Transcriptome profile analysis of flowering molecular processes of early flowering trifoliate orange mutant and the wild-type [Poncirus trifoliata (L.) Raf.] by massively parallel signature sequencing. BMC Genomics 12:63
Zhang J-Z, Li Z-M, Mei L, Yao J-L, Hu C-G (2009) PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuation in expression level. Planta 229:847–859
Zhang JZ, Zhao K, Ai XY, Hu CG (2014) Involvements of PCD and changes in gene expression profile during self-pruning of spring shoots in sweet orange (Citrus sinensis). BMC Genomics 15:892
Acknowledgments
We are grateful to Prof. Jia-Ling Yao for her helpful discussion in this manuscript. This research was supported financially by the National Natural Science Foundation of China (grant nos. 31130046, 31471863, 31360469, 31372046, and 31521092) and Fundamental Research Funds for the Central Universities (Program No. 2662016PY037).
Author contributions
X-YA carried out the yeast two-hybrid, bimolecular fluorescence complementation, and PCR experiments; X-YA and T-JL prepared the plant material and obtained the transgenic plants; J-ZZ and C-GH designed the experiments and the study; X-YA and J-ZZ wrote the paper. All authors discussed the data obtained. All authors reviewed and provided comments upon preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Data archiving statement
The PtFCA1, PtFCA2, and PtFCA3 have been deposited in GenBank under accession nos. KX440390, KX440391, and KX440392, respectively.
Additional information
Communicated by W.-W. Guo
Xiao-Yan Ai and Jin-Zhi Zhang contributed equally to this work.
Electronic supplementary material
Figure S1
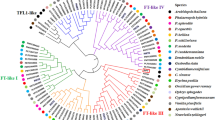
Gene sequences, structures, and phylogenetic relationship of PtFCA. A: Sequence alignment between PtFCA2 and PtFCA3. The gray box represents the sequence differences between PtFCA2 and PtFCA3. B: Alternatively spliced transcripts were validated in precocious trifoliate orange by RT-PCR, M: Marker (DL2000). Lanes 1–4: PtFCA1 in trifoliate orange, 5–7: PtFCA2/3 in precocious trifoliate orange. C: Phylogenic tree of FCA showing evolutionary relationships. The sequences with the highest percent sequence similarity are grouped together. The accession numbers of FCA proteins used in this analysis are HvFCA (FJ188402.2), VvFCA (GU300763.1), RcFCA (XM_002519206.1), GmFCA-like (XM_003529468.1), PsFCAgamma (AY805329.1), MtFCA (XM_003609755.1), FCAalpha (Z82993.1), FCAbeta (Z82991.1), FCAdelta (Z82990.1), FCAgamma (Z82989.1), OsFCA-1 (AY274928.1), OsFCA-2 (AY311344.1), OsFCA-3 (AY311343.1), OsFCA-4 (AY331574.1). (JPEG 4617 kb)
Table S1
List of primer sequences used in this study. (XLS 25 kb)
Rights and permissions
About this article
Cite this article
Ai, XY., Zhang, JZ., Liu, TJ. et al. PtFCA from precocious trifoliate orange is regulated by alternative splicing and affects flowering time and root development in transgenic Arabidopsis . Tree Genetics & Genomes 12, 85 (2016). https://doi.org/10.1007/s11295-016-1035-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1035-6