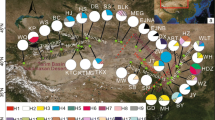
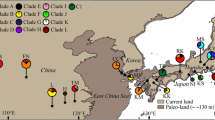
Abstract
Historical geoclimatic events have shaped the distribution patterns and intraspecific divergence of plants. Numerous phylogeographical studies in China have focused on the Qinghai-Tibetan Plateau and surrounding areas due to the complex topography and high species diversity, but the impact of Neogene events and Quaternary climatic change on the flora of subtropical China remains poorly understood. Quercus glauca, a widespread tree of East Asian subtropical evergreen forests, has rich fossil records dating back to the Neogene, and it provides a good model to explore the impact of paleogeoclimate changes on East Asian subtropical forests. We used three chloroplast DNA (cpDNA) intergenic spacer regions and ecological niche modeling (ENM) to analyze the divergence pattern and demographic history of Q. glauca in China and Japan. A total of 33 haplotypes were detected. The phylogenetic analysis revealed two major haplotype lineages (Southwest China vs. Southeast China and East China Sea). The limited dispersal ability of seeds and complex topography resulted in the high total, inter- and intrapopulation haplotype diversity. The fossil-constrained BEAST analysis revealed a lineage diversification in the late Miocene-Pliocene. The formation of complex topography changes since Miocene in east Himalaya and adjacent area might be the key factor that triggered the intraspecific divergence of Q. glauca. Haplotype spatial distribution, ENM, mismatch distribution, and neutrality tests suggest that Q. glauca in Southeast China experiences expansion, and the current distribution in region III might be shaped by southward expansion from regions I and II after last glacial maximum (LGM). Regions I and II were the potential glacial refugia of Q. glauca.






Similar content being viewed by others
References
An ZS, Huang YS, Liu WG, Guo ZT, Steven C, Li L, Warren P, Ning YF, Cai YJ, Zhou WJ, Lin BH, Zhang QL, Cao YN, Qiang XK, Chang H, Wu ZK (2005) Multiple expansions of C-4 plant biomass in East Asia since 7 Ma coupled with strengthened monsoon circulation. Geology 33:705–708
An ZS, Kutzbach JE, Prell WL, Porter SC (2001) Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since Late Miocene times. Nature 411:62–66
Axelrod DI, Al-Shehbaz I, Raven PH (1996) History of the modern flora of China. In: Zhang A, Wu SG (eds) Floristic characteristics and diversity of East Asian plants. China Higher Education Press, Beijing, pp 43–45
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Benton M, Donoghue PCJ, Asher RJ (2009) Calibrating and constraining molecular clocks. In: Hedges SB, Kumar S (eds) The timetree of life. Oxford University Press, Oxford, pp 35–86
Borgardt SJ, Pigg KB (1999) Anatomical and developmental study of petrified Quercus (Fagaceae) fruits from the Middle Miocene, Yakima Canyon, Washington. USA Am J Bot 86(86):307–325
Chen DM, Zhang XX, Kang HZ, Sun X, Yin S, Du HM, Yamanaka N, Gapare W, Wu HX, Liu CJ (2012) Phylogeography of Quercus variabilis based on chloroplast DNA sequence in East Asia: multiple glacial refugia and mainland-migrated island populations. PLoS ONE 7 doi:10.1371/journal.pone.0047268
Chou YW, Thomas PI, Ge XJ, LePage BA, Wang CN (2011) Refugia and phylogeography of Taiwania in East Asia. J Biogeogr 38:1992–2005
Crepet WL, Nixon KC (1989) Extinct transitional Fagaceae from the Oligocene and their phylogenetic implications. Am J Bot 76:1493–1505
Deng M, Li QS, Yang ST, Liu YC, Xu J (2013a) Comparative morphology of leaf epidermis in the genus Lithocarpus and its implication in leaf epidermal feature evolution in Fagaceae. Plant Syst Evol 299:659–681
Deng M, Zhou ZK, Li QS (2013b) Taxonomy and systematics of Quercus subgenus Cyclobalanopsis. Int Oaks 24:48–60
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973
Dumolin S, Demesure B, Petit RJ (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor Appl Genet 91:1253–1256
Elith J, Graham CH, Anderson RP, Dudík M, Ferrier S, Guisan A, Hijmans R, Huettmann F, Leathwick JR, Lehmann A, Li J, Lohmann LG, Loiselle BA, Manion G, Moritz C, Nakamura M, Nakazawa Y, Overton JMM, Peterson AT, Phillips SJ, Richardson K, Scachetti-Pereira R, Schapire RE, Soberón J, Williams S, Wisz MS, Zimmermann NE (2006) Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29:129–151
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fouquet A, Green DM, Waldman B, Bowsher JH, McBride KP, Gemmell NJ (2010) Phylogeography of Leiopelma hochstetteri reveals strong genetic structure and suggests new conservation priorities. Conserv Genet 11:907–919
Frascaria N, Maggia L, Michaud M, Bousquet J (1993) The rbcL gene sequence from chestnut indicates a slow rate of evolution in the Fagaceae. Genome 36:668–671
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gao LM, Moller M, Zhang XM, Hollingsworth ML, Liu J, Mill RR, Gibby M, Li DZ (2007) High variation and strong phylogeographic pattern among cpDNA haplotypes in Taxus wallichiana (Taxaceae) in China and North Vietnam. Mol Ecol 16:4684–4698
Gong W, Chen C, Dobes C, Fu CX, Koch MA (2008a) Phylogeography of a living fossil: Pleistocene glaciations forced Ginkgo biloba L. (Ginkgoaceae) into two refuge areas in China with limited subsequent postglacial expansion. Mol Phylogenet Evol 48:1094–1105
Gong W, Zeng Z, Chen YY, Chen C, Qiu YX, Fu CX (2008b) Glacial refugia of Ginkgo biloba and human impact on its genetic diversity: evidence from chloroplast DNA. J Integr Plant Biol 50:368–374
Govaerts R, Frodin DG (1998) World checklist and bibliography of Fagales (Betulaceae, Corylaceae, Fagaceae and Ticodendraceae). Kew Publishing, London
Graur D, Li WH (2000) Fundamentals of molecular evolution, 2nd edn. Sinauer Associates Inc., Sunderland
Grivet D, Heinze B, Vendramin GG, Petit RJ (2001) Genome walking with consensus primers: application to the large single copy of chloroplast DNA. Mol Ecol Notes 1:345–349
Guo SX (1978) Pliocene flora of western Sichuan. Acta Palaeontol Sin 17:343–349
Guo SX (2011) The late Miocene Bangmai flora from Lincang county of Yunnan, southwestern China. Acta Palaeontol Sin 50:353–408
Guo ZT, Ruddiman WF, Hao QZ, Wu HB, Qiao YS, Zhu RX, Peng SZ, Wei JJ, Yuan BY, Liu TS (2002) Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 416:159–163
Hai LC, Tang LY, Wan HW, Wang SM, Yao SC (2009) Vegetation and human activity in Yuyao (Zhenjiang province) inferred from the sporo-pollen record since the late Pleistocene. Acta Micropalaeontol Sin 26:48–56
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467
Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
He K, Jiang XL (2014) Sky islands of southwest China. I: An overview of phylogeographic patterns. Chin Sci Bull 59:585–597
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linnean Soc 58:247–276
Hsieh YC, Chung JD, Wang CN, Chang CT, Chen CY, Hwang SY (2013) Historical connectivity, contemporary isolation and local adaptation in a widespread but discontinuously distributed species endemic to Taiwan, Rhododendron oldhamii (Ericaceae). Heredity 111:147–156
Huang CC, Chang YT, Bartholomew B (1999) Fagaceae. In: Wu CY, Raven PH (eds) Flora of China, vol 4. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis, pp 380–400
Huang SSF, Hwang SY, Lin TP (2002) Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and East Asia. Mol Ecol 11:2349–2358
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Jia H, Sun BN, Li XC, Xiao L, Wu JY (2009) Microstructures of one species of Quercus from the Neogene in Eastern Zhejiang and its palaeoenvironmental indication. Earth Sci Front 16:79–90
Jimenez-Valverde A, Lobo JM (2007) Threshold criteria for conversion of probability of species presence to either-or presence-absence. Acta Oecol 31:361–369
Kelchner SA, Thomas MA (2007) Model use in phylogenetics: nine key questions. Trends Ecol Evol 22:87–94
Kono Y, Chung KF, Chen CH, Hoshi Y, Setoguchi H, Chou CH, Oginuma K, Peng CI (2012) Intraspecific karyotypic polymorphism is highly concordant with allozyme variation in Lysimachia mauritiana (Primulaceae: Myrsinoideae) in Taiwan: implications for the colonization history and dispersal patterns of coastal plants. Ann Bot 110:1119–1135
Kvacek Z, Teodoridis V (2007) Tertiary macrofloras of the Bohemian Massif: a review with correlations within Boreal and Central Europe. Bull Geosci 82:383–408
Kvacek Z, Walther H (1989) Paleobotanical studies in Fagaceae of the European Tertiary. Plant Syst Evol 162:213–229
Lei M, Wang Q, Wu ZJ, López-Pujol J, Li DZ, Zhang ZY (2012) Molecular phylogeography of Fagus engleriana (Fagaceae) in subtropical China: limited admixture among multiple refugia. Tree Genet Genome 8:1203–1212
Li Y, Yan HF, Ge XJ (2012) Phylogeographic analysis and environmental niche modeling of widespread shrub Rhododendron simsii in China reveals multiple glacial refugia during the last glacial maximum. J Syst Evol 50:362–373
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lin TP, Chuang WJ, Huang SSF, Hwang SY (2003) Evidence for the existence of some dissociation in an otherwise strong linkage disequilibrium between mitochondrial and chloroplastic genomes in Cyclobalanopsis glauca. Mol Ecol 12:2661–2668
Liu JQ, Sun YS, Ge XJ, Gao LM, Qiu YX (2012) Phylogeographic studies of plants in China: advances in the past and directions in the future. J Syst Evol 50:267–275
Liu L, Hao ZZ, Liu YY, Wei XX, Cun YZ, Wang XQ (2014) Phylogeography of Pinus armandii and its relatives: heterogeneous contributions of geography and climate changes to the genetic differentiation and diversification of Chinese white pines. Plos One 9(1):e85920. doi:10.1371/journal.pone.0085920
Lopez-Pujol J, Zhang FM, Sun HQ, Ying TS, Ge S (2011) Centres of plant endemism in China: places for survival or for speciation? J Biogeogr 38:1267–1280
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annu Rev Ecol Syst 15:65–95
McLaughlin BC, Zavaleta ES (2012) Predicting species responses to climate change: demography and climate microrefugia in California valley oak (Quercus lobata). Glob Chang Biol 18:2301–2312
Médail F, Diadema K (2009) Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J Biogeogr 36:1333–1345
Miao YF, Herrmann M, Wu FL, Yan XL, Yang SL (2012) What controlled Mid-Late Miocene long-term aridification in central Asia?—global cooling or Tibetan plateau uplift: a review. Earth-Sci Rev 112:155–172
Morris AB, Graham CH, Soltis DE, Soltis PS (2010) Reassessment of phylogeographical structure in an eastern North American tree using Monmonier’s algorithm and ecological niche modelling. J Biogeogr 37:1657–1667
Mosca E, Eckert AJ, Di Pierro EA, Rocchini D, La Porta N, Belletti P, Neale DB (2012) The geographical and environmental determinants of genetic diversity for four alpine conifers of the European Alps. Mol Ecol 21:5530–5545
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Nakamura K, Denda T, Kokubugata G, Suwa R, Yang TYA, Peng C-I, Yokota M (2010) Phylogeography of Ophiorrhiza japonica (Rubiaceae) in continental islands, the Ryukyu Archipelago, Japan. J Biogeogr 37:1907–1918
Nei M (1975) Molecular population genetics and evolution. North Holland, Amsterdam
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A 76:5269–5273
Ni J, Song YC (1997) Relationships between geographical distribution of Cyclobalanopsis glauca and climate in China. Acta Bot Sin 39:451–460
Oh SH, Manos PS (2008) Molecular phylogenetics and cupule evolution in Fagaceae as inferred from nuclear CRABS CLAW sequences. Taxon 57:434–451
Pages RDM, Holmes E (1998) Molecular evolution, a phylogenetic approach. Blackwell, Oxford
Peakall ROD, Smouse PE (2006) genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Phillips SJ, Dudik M, Schapire RE (2004) A maximum entropy approach to species distribution modeling. In: Proceedings of the twenty-first international conference on Machine learning. Banff, Canada, 655–662
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Qi XS, Chen C, Comes HP, Sakaguchi S, Liu YH, Tanaka N, Sakio H, Qiu YX (2012) Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol 196:617–630
Qian H, Ricklefs RE (2000) Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407:180–182
Qiu YX, Fu CX, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol 59:225–244
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Bio Evol 9:552–569
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Rousset F, Raymond M (1995) Testing heterozygote excess and deficiency. Genetics 140:1413–1419
Sauquet H, Ho SY, Gandolfo MA, Jordan GJ, Wilf P, Cantrill DJ, Bayly MJ, Bromham L, Brown GK, Carpenter RJ, Lee DM, Murphy DJ, Sniderman JM, Udovicic F (2012) Testing the impact of calibration on molecular divergence times using a fossil-rich group: the case of Nothofagus (Fagales). Syst Biol 61:289–313
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166
Shi SQ, Yuan DX, Luo LD, Zhao ZY, Hao XD (2012) Sporopollen records and climate changes since 35,000 aBP in Hongya, Sichuan Province. Carsol Sin 31:121–130
Shi MM, Michalski SG, Welk E, Chen XY, Durka W (2014) Phylogeography of a widespread Asian subtropical tree: genetic east–west differentiation and climate envelope modelling suggest multiple glacial refugia. J Biogeogr 41:1710–1720
Shih FL, Cheng YP, Hwang SY, Lin TP (2006a) Partial concordance between nuclear and organelle DNA in revealing the genetic divergence among Quercus glauca (Fagaceae) populations in Taiwan. Int J Plant Sci 167:863–872
Shih HT, Hung HC, Schubart CD, Chen CLA, Chang HW (2006b) Intraspecific genetic diversity of the endemic freshwater crab Candidiopotamon rathbunae (Decapoda, Brachyura, Potamidae) reflects five million years of the geological history of Taiwan. J Biogeogr 33:980–989
Simeone MC, Piredda R, Papini A, Vessella F, Schirone B (2013) Application of plastid and nuclear markers to DNA barcoding of Euro-Mediterranean oaks (Quercus, Fagaceae): problems, prospects and phylogenetic implications. Bot J Linn Soc 172:478–499
Song SY, Krajewska K, Wang YF (2000) The first occurrence of the Quercus section Cerris Spach fruits in the Miocene of China. Acta Palaeobotanica 40:153–163
Sork VL, Davis FW, Westfall R, Flint A, Ikegami M, Wang H, Grivet D (2010) Gene movement and genetic association with regional climate gradients in California valley oak (Quercus lobata Née) in the face of climate change. Mol Ecol 19:3806–3823
Sun Y, Hu HQ, Huang HW, Vargas-Mendoza CF (2014) Chloroplast diversity and population differentiation of Castanopsis fargesii (Fagaceae): a dominant tree species in evergreen broad-leaved forest of subtropical China. Tree Genetics & Genomes 10:1531–1539
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Tang ZH, Ding Z, White PD, Dong X, Ji J, Jiang H, Luo P, Wang X (2011) Late Cenozoic central Asian drying inferred from a palynological record from the northern Tian Shan. Earth Planet Sci Lett 302:439–447
Tiffney BH, Manchester SR (2001) The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. Int J Plant Sci 162:S3–S17
Tribsch A (2004) Areas of endemism of vascular plants in the Eastern Alps in relation to Pleistocene glaciation. J Biogeogr 31:747–760
Tribsch A, Schonswetter P (2003) Patterns of endemism and comparative phylogeography confirm palaeoenvironmental evidence for Pleistocene refugia in the Eastern Alps. Taxon 52:477–497
Wang J, Gao P, Kang M, Lowe AJ, Huang H (2009) Refugia within refugia: the case study of a canopy tree (Eurycorymbus cavaleriei) in subtropical China. J Biogeogr 36:2156–2164
Whitlock BA, Hale AM, Groff PA (2010) Intraspecific inversions pose a challenge for the trnH-psbA plant DNA barcode. PLoS One 55(7):e11533. doi:10.1371/journal.pone.0011533
Wu ZY (1980) China’s vegetation. Science Press, Beijing
Xiao J, Lü H, Zhou W, Zhao Z, Hao R (2007) Evolution of vegetation and climate since the last glacial maximum recorded at Dahu peat site, South China. Sci China Ser D Earth Sci 50:1209–1217
Xiao ZS, Zhang ZB, Wang YS (2005) Effects of seed size on dispersal distance in five rodent-dispersed fagaceous species. Acta Oecol 28:221–229
Yan HF, Zhang CY, Wang FY, Hu CM, Ge XJ, Hao G (2012) Population expanding with the phalanx model and lineages split by environmental heterogeneity: a case study of Primula obconica in subtropical China. PLoS One 7(9):e41315. doi:10.1371/journal.pone.0041315
Ying TS (2001) Species diversity and distribution patterns of seed plants in China. Biodivers Sci 9:393–398
Ying TS, Zhang YL, Bufford DE (1993) The endemic genera of seed plants of China. Science Press, Beijing
Yue LL, Chen G, Sun WB, Sun H (2012) Phylogeography of Buddleja crispa (Buddlejaceae) and its correlation with drainage system evolution in southwestern China. Am J Bot 99:1726–1735
Zhang DR, Chen MY, Murphy RW, Che J, Pang JF, Hu JS, Luo J, Wu SJ, Ye H, Zhang YP (2010) Genealogy and palaeodrainage basins in Yunnan Province: phylogeography of the Yunnan spiny frog, Nanorana yunnanensis (Dicroglossidae). Mol Ecol 19:3406–3420
Zhang ZY, Wu R, Wang Q, Zhang ZR, Lopez-Pujol J, Fan DM, Li DZ (2013) Comparative phylogeography of two sympatric beeches in subtropical China: species-specific geographic mosaic of lineages. Ecol Evol 3:4461–4472
Acknowledgments
We thank Dr. Yun-Juan Zuo for the help on data analysis and the anonymous reviewers for their insightful and valuable suggestions/comments that further improved the manuscript. We are grateful to Dr. Tao Su, Ms. Jin-jin Hu (Xishuangbanna Tropical Botanical Garden, CAS), Dr. Dai-Ke Tian, Dr. Yue-hong Yan, Qi Tian, Bin-jie Ge, Dr. Yan-Chun Liu, Xiang-peng Li, Yan-bo Huang, Tao Sang, Hui Shang, and Chun Li (Shanghai Chenshan Plant Science Research Center, CAS) for their help in the sample collection. This work was supported by grants from the National Natural Science Foundation of China (31100154, 31270267, 31110103911), the Shanghai Municipal Administration of Forestation and City Appearances (F112419), Science and Technology Basic Work (2013FY112100), and State Key Laboratory of Systematic and Evolutionary Botany (grant no. LSEB2011-02).
Author contributions
Min Deng conceived and designed the experiments; Jin Xu performed the experiments; Jin Xu and Xiao-Long Jiang analyzed the data; Min Deng and Yi-Gang Song were responsible for field collections and specimen identification; Murphy Westwood and Roy Turkington contributed analysis tools and revised the manuscript; Min Deng, Jin Xu, and Xiao-Long Jiang wrote the paper.
Data Archiving Statement
Sequence data obtained in this study has been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The accession numbers were KM210592-KM210622, KM210623-KM210650, and KM210651-KM210668 for atpI-atpH, psbA-trnH, and trnT-trnL, respectively.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: A. Kremer
Jin Xu and Min Deng contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 37 kb)
Fig. S1
Correlation between genetic distances (F ST) and geographical distances (kilometers) separating each pairwise combination of populations within Q. glauca (P = 0.097). (GIF 56 kb)
Fig. S2
Chronograms of haplotypes of Q. glauca, with Q. variabilis as outgroup based on a Bayesian analysis. Calibration points are marked with black solid circled. The numbers at the nodes suggested the TMRCA of the lineages. (GIF 17 kb)
Rights and permissions
About this article
Cite this article
Xu, J., Deng, M., Jiang, XL. et al. Phylogeography of Quercus glauca (Fagaceae), a dominant tree of East Asian subtropical evergreen forests, based on three chloroplast DNA interspace sequences. Tree Genetics & Genomes 11, 805 (2015). https://doi.org/10.1007/s11295-014-0805-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-014-0805-2