Abstract
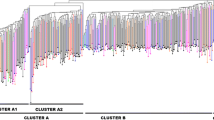
Loquat (Eriobotrya japonica (Thunb.) Lindl., Maloideae, Rosaceae) is a subtropical evergreen fruit tree indigenous of China, where the center of origin of the species is located. Loquat is grown in all subtropical areas and was introduced in the Mediterranean basin in late eighteenth century. In Europe, the largest germplasm bank is located at Instituto Valenciano de Investigaciones Agrarias (IVIA; Valencia, Spain). Thirteen microsatellites and a conserved region of S-allele were used to assess the genetic diversity of 102 accessions of the IVIA collection. A total of 38 SSR alleles and 11 putative S-alleles were used to study the genetic structure of the loquat germplasm bank using the STRUCTURE software, Factorial Correspondence Analysis (FCA), and unweighted pair-group method (UPGMA) cluster analyses. The total diversity was H T = 0.5682, the genetic differentiation G ST = 0.1660, and the standardized G ST reached a much higher value of G′ST = 0.4948. The Evanno’s test indicated that the most informative number of populations was five, with accessions distributed according to their geographic origin in two, one, and two groups of Spanish, Italo-Spanish, and non-European origin, respectively. Knowledge of the substructure and diversity of the IVIA loquat collection and the self-incompatibility genotype data will allow us to select and incorporate useful materials into the loquat breeding program.



Similar content being viewed by others
References
Atayde MO, Fornazier MJ, Da Costa AN, Nunes FAR (1992) Evaluation of loquat cultivars for the Serrana region of Espiritu Santo. Hort Abstr 62:7821
Badenes ML, Martínez-Calvo J, Llacer G (2000) Analysis of a germplasm collection of loquat (Eriobotrya japonica Lindl.). Euphytica 114:187–194
Badenes ML, Canyamás TP, Romero C, Giordani E, Martínez-Calvo J, Llácer G (2004) Characterization of underutilized fruits by molecular markers, loquat a case of study. Genet Resour Crop Ev 51:335–341
Badenes ML, Lin S, Yang X, Liu C, Huang X (2009) Loquat (Eriobotrya Lindl.). In: Folta KV, Gardiner SE (eds) Genetics and genomics of Rosaceae. Springer, New York, pp 525–538
Badenes ML, Janick J, Lin S, Zhang Z, Liang GL, Wang W (2013) Breeding loquat. Plant Breed Rev 37:259–296
Badenes ML, Blasco M, Naval MM, Soler E (2014) Integrating Biotechnology in loquat breeding. IV International Symposium of loquat. May 12–15, Palermo, Italy. Acta Horticulturae (in press)
Baratta B, Campisi G, Raimondo A (1995) Miglioramento genetico del nespolo del Giappone (Eriobotrya japonica Linde) cv. Marchetto. Riv Frutt Ortoflor 57(1):27–32
Barkley NA, Roose ML, Krueger RR, Federici CT (2006) Assessing genetic diversity and population structure in a citrus germplasm utilizing simple sequence repeat markers (SSRs). Theor Appl Genet 112:1519–1531
Belkhir K, Catric V, Bonhomme F (2000) GENETIX 4.05, Logiciel Sous Windows™ pour la Génétique des Populations. Montpellier: Laboratoire Génome, Université Montpellier. http://kimura.univ-montp2.fr/genetix [last accessed 01.06.2014]
Breton C, Tersac M, Bervillé A (2006) Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. J Biogeogr 33:1916–1928
Burle ML, Fonseca JR, Kauni JA, Gepts P (2010) Microsatellite diversity and genetic structure among bean (Phaseolus vulgaris L.) landcraces in Brazil, a secondary center of diversity. Theor Appl Genet 121:801–8013
Cai LH (2000) Allozyme analysis of genetic diversity interspecific relationship and cultivar identification in genus Eriobotrya. Dissertation, Huazhong Agricultural University
Couviour F, Faure S, Poupard B, Flodrops Y, Dubreuil P, Praud S (2011) Analysis of genetic structure in a panel of elite wheat varieties and relevance for association mapping. Theor Appl Genet 123:715–727
Ding CK, Chen QF, Sun TL, Xia QZ, Zhu DW (1995) Germplasm resources and breeding of Eryobotria japonica Lindl. in China. Acta Hort 403:121–126
Doyle JJ, Doyle JL (1987) A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martínez-Zapater J, Zyprian E, Moreira FM, Grando MS (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol 13:39
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Frascaroli STA, Melchinger AE (2013) Genetic diversity analysis of elite European maize (Zea mays L.) inbred lines using AFLP, SSR, and SNP markers reveals ascertainment bias for a subset of SNPs. Theor Appl Genet 126:133–141
Gianfranceschi L, Seglias N, Tarchini R (1998) Simple sequence repeats for the genetic analysis of apple. Theor Genet 96:1069–1076
Gisbert AD, Martinez-Calvo J, Llácer G, Romero C, Badenes ML (2009a) Genetic diversity evaluation of a loquat (Eriobotrya japonica (Thun)Lindl) germplasm collection by SSR and S-allele fragments. Euphytica 168(1):121–134
Gisbert AD, López-Capuz I, Soriano JM, Romero C, Llácer G, Badenes ML (2009b) Development of microsatellite markers of loquat (Eriobotrya japonica (Thunb.) Lindl.). Mol Ecol Notes 9(3):803–805
Gisbert AD, Martínez-Calvo J, Llácer G, Badenes ML, Romero C (2009c) Development of two loquat (Eriobotrya japonica (Thunb.) Lindl.) linkage maps based on AFLP and SSR markers from different Rosaceae species. Mol Breed 23:523–538
Hurtado M, Vilanova S, Plazas M, Gramazio P, Fonseka HH et al (2012) Diversity and Relationships of Eggplants from Three Geographically Distant Secondary Centers of Diversity. PLoS ONE 7(7):e41748
Langella O (2002) Populations 1.2.32: a population genetic software. CNRS UPR9034. http://bioinformatics.org/~tryphon/populations/. Accessed 25 Feb 2014
Lin SQ (2004) Plant material of loquat in Asian countries. First International Symposium on loquat. Options Méditérr 58:41–44
Lin SQ, Sharpe RH, Janick J (1999) Loquat: botany and horticulture. Hortic Rev Am Soc Hortic Sci 23:233–276
Lin SQ, Yang XH, Liu CM et al (2004) Natural geographical distribution of genus Eriobotrya plants in China. Acta Hort 31(5):569–573
Llácer G, Badenes ML, Martínez-Calvo J (2002) El níspero: estudios sobre el material vegetal en los Países Mediterráneos. Agrícola Vergel 250:552–557
Martínez-Calvo J, Badenes ML, Llácer G (2000) Descripción de variedades de níspero japonés. Publicaciones de la Consellería de Agricultura, Pesca y Alimentación. Serie Divulgación Técnica 47, p 119
Martínez-Calvo J, Badenes ML, Llácer G (2006) Descripción de nuevas variedades de níspero japonés del banco de germoplasma del IVIA. Monografías INIA, Serie Agrícola 21:73
Martínez-Calvo J, Badenes ML, Llácer G (2008) Descripción de 35 nuevas variedades de níspero japonés del banco de germoplasma del IVIA. Monografías INIA, Serie Agrícola 24:73
Mohammadi SA, Prasanna BM (2003) Analysis of Genetic Divesity in Crop Plants. Salient Statistical Tools and considerations. Crop Sci 43:1235–1248
Morton JF (1987) Loquat. In: Fruits of warm climates. Creative Resource Systems, Inc., Winterville, p 103–108
Naval MM, Zuriaga E, Pecchioli S, Llácer G, Giordani E, Badenes ML (2010) Analysis of genetic diversity among persimmon cultivars using microsatellite markers. Tree Genet Genomes 6:677–687
Nei M (1972) Genetic distance among populations. Am Nat 106:283–292
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70:3321–3323
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raspé O, Kohn JR (2002) S-allele diversity in Sorbus aucuparia and Crataegus monogyna (Rosaceae: Maloideae). Heredity 88:458–465
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Soriano JM, Romero C, Vilanova S, Llácer G, Badenes ML (2005) Genetic diversity of loquat (Eriobotrya japonica (Thunb) Lind) assessed by SSR markers. Genome 48:108–114
Weir BS (1990) Genetic data analysis. Methods for discrete genetic data. Sinauer Associates, Suderland
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395–420
Yang XH, Glakpe K, Lin SQ, Hu YL, He YH, Yuanshi JR, Liu YX, Hu GB, Liu CM (2005) Taxa of plants of genus Eriobotrya around the world and native of Southeastern Asia. J Fruit Sci 22(1):55–60
Zhang HZ, Peng SA, Cai LH, Fang DQ (1993) The germplasm resources of the genus Eriobotrya with special reference on the origin of E. japonica Lindl. Plant Breed Abstr 63:772
Zheng SQ (2007) Achievement and prospect of loquat breeding in China. Acta Hort 750:85
Acknowledgments
The authors acknowledge Prof Janick from revision and edition of the manuscript. This research was funded by grant RF 2010-0003-00-00 from the National Agency “Instituto Nacional de Investigaciones Agrarias” (INIA). MBV was funded by a fellowship from INIA.
Data Archiving Statement
The SSR markers used in this study were submitted to the GeneBank; the accession numbers are indicated in Table 2 and reference herein. The results obtained were not susceptible of submission to public databases.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Chagné
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Alleles obtained of the 102 accessions using 13 microsatellite and a conserved region of S-allele (DOC 448 kb)
Rights and permissions
About this article
Cite this article
Blasco, M., Naval, M.d.M., Zuriaga, E. et al. Genetic variation and diversity among loquat accessions. Tree Genetics & Genomes 10, 1387–1398 (2014). https://doi.org/10.1007/s11295-014-0768-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0768-3