Abstract
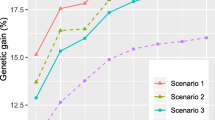
When selecting a clonal mixture for clonal forestry, a common practice is to specify a minimum acceptable genetic diversity for the mixture, and under that constraint, to maximize its genetic gain. Three selection methods—combined index selection (CIS), clonal mean selection (GMS), and family plus within-family clonal selection (FWFGS)—together with various restrictions on family contributions (family restrictions) were used to estimate gain at a given effective population size (N e ) of clonal mixtures selected from a clonally replicated genetic test of white spruce. The designated target trait for improvement was individual tree volume at age 14 after planting. Regardless of selection method, all genetic gains were >30% at given N e from five to 20, suggesting that implementing clonal forestry was a very effective deployment strategy. Genetic gain at a specified N e could be enhanced by using appropriate selection methods: CIS was the most effective, followed by GMS and FWFGS. With an N e of 15, CIS resulted in an average gain of 43.2%, which corresponded to an increase by 8.1% and 17.3% relative to GMS and FWFGS, respectively. Imposing family restriction increased gain at an N e . Compared with the respective unrestricted selection, family restrictions increase genetic gain by 3.3%, 7.7%, and 54% for CIS, GMS, and FWFGS, respectively. The optimal family restriction level for each selection method varied with the specified N e .



Similar content being viewed by others
References
Alberta Sustainable Resource Development (2003) Standards for tree improvement in Alberta. http://www3.gov.ab.ca/srd/forests/fmd/manuals/index.html. Accessed 28 May 2008
Baltunis BS, Huber DA, White TL, Goldfarb B, Stelzer HE (2007) Genetic gain from selection for rooting ability and early growth in vegetatively propagated clones of loblolly pine. Tree Genet Genom 3:227–238
British Columbia Ministry of Forests and Range (2004) Chief forester's standards for seed use. http://www.for.gov.bc.ca/code/cfstandards/html/. Accessed 18 May 2008
Burdon RD, Shelbourne CJA (1974) The use of vegetative propagules for obtaining genetic information. N Z J For Sci 4:418–428
Carson SD, Garcia O, Hayes JD (1999) Realized gain and prediction of yield with genetically improved Pinus radiata in New Zealand. For Sci 45:186–200
Cotterill PP, James JW (1981) Optimizing two-stage independent culling selection in tree and animal breeding. Theor Appl Genet 59:67–72
Cyr DR, Klimaszewska K (2002) Conifer somatic embryogenesis: II. Applications. Dendrobiology 48:41–49
Danusevičius D, Lindgren D (2008) Strategies for optimal development of related clones into seed orchards. Silvae Genet 57:119–127
Falconer DS, MacKay TFC (1996) Introduction to quantitative genetics. Longman Group Ltd., London, p 464
Foster GS, Shaw DV (1988) Using clonal replicates to explore genetic variation in a perennial plant species. Theor Appl Genet 76:788–794
Funda T, Listibůrek M, Lachout P, Klápště J, EI-Kassaby YA (2009) Optimization of combined genetic gain and diversity for collection and deployment of orchard crops. Tree Genet Genom 5:583–593
Gilmour AR, Gogel BJ, Cullis BR, Welham SJ, Thompson R (2002) ASReml user guide release 1.0. VSN interactional Ltd, Hemel Hempstead, HP11ES, p 267
Honer TG, Ker MF, Alemdag IS (1983) Metric timber tables for the commercial tree species of central and eastern Canada. Nat Res Can Can For Serv—Atl For Ctr Inf Rep M-X-140
Isik F, Li B, Frampton J, Goldfarb B (2004) Efficiency of seedlings and rooted cuttings for testing and selection in Pinus taeda. For Sci 50:44–53
Isik F, Goldfarb B, Lebude A, Li B, McKeand S (2005) Predicted genetic gains and testing efficiency from two loblolly pine clonal trials. Can J For Res 35:1754–1766
King JN, Johnson GR (1993) Monte Carlo simulation models of breeding-population advancement. Silvae Genet 42:68–78
Klimaszewska KK, Trontin JF, Becwar MR, Devillard C, Park YS, Lelu-Walter MA (2007) Recent progress in somatic embryogenesis of four Pinus spp. Tree For Sci Biotech 1:11–25
Kumar S (2006) Correlation between clonal means and open-pollinated seedling progeny means and its implications for radiata pine breeding strategy. Can J For Res 36:1968–1975
Libby WJ (1982) What is a safe number of clones per plantation? In: Heybroek HM, Stephan BR, von Weisenberg K (eds) Resistance to disease and pests in forest trees: proceedings of the third international workshop on the genetics of host-parasite interactions. Centre for Agricultural Publishing and Document, Wageningen, pp 342–360
Lindgren D (1993) The population biology of clonal deployment. In: Ahuja MR, Libby WJ (eds) Clonal forestry I: genetics and biotechnology. Springer-Verlag, Berlin, pp 34–49
Lindgren D, Matheson AC (1986) An algorithm for increasing the genetic quality of seed from seed orchards by using the better clones in higher proportions. Silvae Genet 35:173–175
Lindgren D, Mullin TJ (1997) Balancing gain and relatedness in selection. Silvae Genet 46:124–129
Lindgren D, Mullin TJ (1998) Relatedness and status number in seed orchard crops. Can J For Res 28:122–129
Lindgren D, Danusevicius D, Rosvall O (2009) Unequal development of clones to seed orchards by considering genetic gain, relatedness and gene diversity. Forestry 82:17–28
Littell RC, Stroup WS, Freund RJ (2002) SAS for linear models, 4th edn. SAS Institute Inc, Cary, NC, p 466
Matheson AC, Lindgren D (1985) Gains from the clonal and the clonal seed orchard options compared for tree breeding programs. Theor Appl Genet 71:242–249
McInnis B, Tosh K (2004) Genetic gains from 20 years of cooperative tree improvement in New Brunswick. For Chron 80:127–133
Mullin TJ, Park YS (1992) Estimating genetic gains from alternative breeding strategies for clonal forestry. Can J For Res 22:14–23
Mullin TJ, Park YS (1994) Genetic parameters and age-age correlations in a clonally replicated test of black spruce after 10 years. Can J For Res 24:2330–2341
New Brunswick Tree Improvement Council (NBTIC) (1989) White spruce progeny test. Est Rep No 30. Fredericton, NB
Park YS (2002) Implication of conifer somatic embryogenesis in clonal forestry: technical requirements and deployment considerations. Ann For Sci 59:651–656
Park YS, Barrett JD, Bonga JM (1998a) Application of somatic embryogenesis in high-value clonal forestry: deployment, genetic control, and stability of cryopreserved clones. In Vitro Cell Dev Biol Plant 34:231–239
Park YS, Bonga JM, Mullin TJ (1998b) Clonal forestry. In: Mandal AK, Gibson GL (eds) Forest genetics and tree breeding. CBS Publishers & Distributors, pp 143–167
Roberds JH, Bishir JW (1997) Risk analyses in clonal forestry. Can J For Res 27:425–432
Statistical Analysis System (SAS) (1990) SAS procedures guides. Version 6, 3rd edn. SAS Institute Inc, Cary, NC
Statistical Analysis System (SAS) (1999) SAS/IML user's guide, version 8. SAS Institute Inc, Cary, NC, p 846
Stoehr M, Webber J, Woods J (2004) Protocol for rating seed orchard seedlots in British Columbia: quantifying genetic gain and diversity. Forestry 77:297–303
Sutton B (2002) Commercial delivery of genetic improvement to conifer plantations using somatic embryogenesis. Ann For Sci 59:657–661
Sutton BCS, Grossnickle SC, Roberts DR, Russell JH, Kiss GK (1993) Somatic embryogenesis and tree improvement in interior spruce. J For 91:34–38
Wei RP (1995a) Response to selection with restrictions with considering effective family number. Hereditas 123:53–59
Wei RP (1995b) Optimal restricted phenotypic selection. Theor Appl Genet 91:289–394
Wei RP, Lindgren D, Yeh FC (1997) Expected gain and status number following restricted individual and combined-index selection. Genome 40:1–8
Weng YH, Park YS, Krasowski MJ, Tosh KJ, Adams G (2008a) Partitioning of genetic variance and selection efficiency for alternative vegetative deployment strategies for white spruce in eastern Canada. Tree Genet Genom 4:809–819
Weng YH, Tosh K, Adam G, Fullarton MS, Norfork C, Park YS (2008b) Realized genetic gains observed in a first generation seedling seed orchard for jack pine in New Brunswick, Canada. New For 36:285–298
White TL, Hodge GR (1989) Predicting breeding values with applications in forest tree improvement. Kluwer Academic Publishers, Dordrecht, p 367
White TL, Adams WT, Neale DB (2007) Forest genetics. CABI Publishing, Cambridge, MA, p 682
Wu HX, Matheson AC (2004) General and specific combining ability from partial diallels of radiata pine: implications for utility of SCA in breeding and deployment populations. Theor Appl Genet 108:1503–1512
Xiang B, Li B (2003) Best linear unbiased prediction of clonal breeding values and genetic values from full-sib mating designs. Can J For Res 33:2036–2043
Zheng YQ, Lindgren D, Rosvall O, Westin J (1997) Combining genetic gain and diversity by considering average coancestry in clonal selection of Norway spruce. Theor Appl Genet 95:1312–1319
Acknowledgments
We would like to thank Drs. John Russell and Judy Loo for their useful comments/suggestions on an earlier version of the paper. The authors are grateful to Dr. Dag Lindgren for helpful discussion on “optimal selection” and providing us a procedure for its calculation. Valuable comments/suggestions from Dr. R. Burdon and two reviewers are also greatly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Burdon
Rights and permissions
About this article
Cite this article
Weng, Y., Park, Y.S. & Krasowski, M.J. Managing genetic gain and diversity in clonal deployment of white spruce in New Brunswick, Canada. Tree Genetics & Genomes 6, 367–376 (2010). https://doi.org/10.1007/s11295-009-0255-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-009-0255-4