Abstract
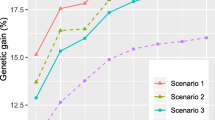
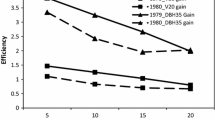
Genetic variances and selection efficiencies for growth traits of white spruce (Picea glauca (Moench) Voss) were estimated from clonally replicated full-sib progeny tests established both in nursery and field environments in New Brunswick, Canada. The available data included heights at 4, 5, and 6 years in the nursery test; height at 9 years, height, DBH, and volume at 14 years in the field test. Estimated variance components were interpreted according to an additive-dominance-epistasis model. For heights in the nursery test, while both non-additive and additive variances were important sources of genetic variation, the former decreased but the latter increased with age; among the non-additive genetic variance, the epistatic variance was much more important than the dominance variance. Different from the nursery traits, for traits in the field test, additive variance accounted for an average of 81% of the total genetic variance, whereas dominance variance explained most of the remaining genetic variance. Genetic parameters and selection efficiencies for three vegetative deployment strategies: deploying half-sib families (VD_FAMHS), full-sib families (VD_FAMFS), and multi-varietal forestry (MVF), were compared. Heritability estimates were moderate for VD_FAMHS and VD_FAMFS (0.61–0.72), high for MVF (>0.82) for the nursery heights, and high (>0.79) for the field traits for all strategies. Genetic correlations of volume at age 14 in the field test, the target trait for improvement, were strong (>0.85) with other field traits. Genetic correlations of VOL14 with the nursery heights were also strong (>0.71) at the half-sib and full-sib family levels, but were only moderate (>0.59) for MVF. Overall, practicing MVF is the most effective deployment strategy, yielding the highest genetic gains, followed by VD_FAMFS and VD_FAMHS, regardless of traits and selection methods. Furthermore, early selections for HT9 or for HT4–HT6 were very encouraging, resulting in higher gain in volume at age 14 on a per year basis.


Similar content being viewed by others
References
Balocchi CE, Bridgwater FE, Zobel BJ, Jahromi SI (1993) Age trends in genetic parameters for tree height in a non-selected population of loblolly pine. For Sci 39:231–251
Baltunis BS, Huber DA, White TL, Goldfarb B, Stelzer HE (2007a) Genetic analysis of early field growth of loblolly pine clones and seedlings from the same full-sib families. Can J For Res 37:195–205
Baltunis BS, Huber DA, White TL, Goldfarb B, Stelzer HE (2007b) Genetic gain from selection for rooting ability and early growth in vegetatively propagated clones of loblolly pine. Tree Genetics & Genomes 3:227–238
Bentzer BG (1993) Strategies for clonal forestry with Norway spruce. In: Ahuja MR, Libby WJ (eds) Clonal forestry II: Conservation and application. Spring, Berlin, pp 120–138
Bentzer BG, Foster GS, Hellberg AR, Podzorski AC (1988) Genotype × environment interaction in Norway spruce involving three levels of genetic control: seed source, clone mixture, and clone. Can J For Res 18:1172–1181
Burdon RD, Shelbourne CJA (1974) The use of vegetative propagules for obtaining genetic information. NZ J For Sci 4:418–428
Callister AN, Collins SL (2008) Genetic parameter estimates in a clonally replicated progeny test of teak (Tectona grandis Linn. F.). Tree Genetics & Genomes 4(2):237–245
Cannell MGR, Sheppard LJ, Cahalan CM (1988) C effects and second generation clone performance in Picea sitchensis and Pinus contorta. Silvae Genet 37:15–19
Carson SD (1990) Genotype × environment interaction and optimal number of progeny tests for improving Pinus radiata in New Zealand. NZ J For Sci 21:32–49
Costa e Silva J, Borralho NMG, Potts BM (2004) Additive and non-additive genetic parameters from clonally replicated and seedling progenies of Eucalyptus globulus. Theor Appl Genet 108:1113–1119
Cotterill PP, Dean CA (1988) Changes in genetic control of growth of radiata pine to 16 years and efficiencies of early selection. Silvae Genet 37:138–146
Dieters MJ, White TL, Littell RC, Hodge GR (1995) Application of approximate variances of variance components and their ratios in genetic tests. Theor Appl Genet 91:15–24
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics, 4th edn. Prentice Hall, Harlow, p 464
Foster GS, Shaw DV (1988) Using clonal replicates to explore genetic variation in a perennial plant species. Theor Appl Genet 76:788–794
Franklin EC (1979) Model relating levels of genetic variance to stand development of four North American conifers. Silvae Genet 28:207–212
Gilmour AR, Gogel BJ, Cullis BR, Welham SJ, Thompson R (2002) ASReml user guide release 1.0. VSN International, Hemel Hempstead, p 267
Gleed JA (1993) Development of plantlings and stecklings of radiata pine. In: Ahuja MR, Libby WJ (eds) Clonal forestry II: Conservation and application. Spring, Berlin, pp 149–157
Hayman BI (1963) Notes on diallel cross theory. In: Statistical genetics and plant breeding. NAS–NRC, pp 571–578
Högberg KA, Karlsson B (1998) Nursery selection of Picea abies clones and effects in field trials. Scand J For Res 13:12–20
Honer TG, Ker MF, Alemdag IS (1983) Metric timber tables for the commercial tree species of central and eastern Canada. NRCan, CFS–AFC, Fredericton, New Brunswick, Inf. Rep. M-X-140
Huehn M, Kleinschmit J, Svolba J (1987) Some experimental results concerning age dependency of different components of variance in testing Norway spruce (Picea abies (L.) Harst.) clones. Silvae Genet 36:68–71
Hühn M (1986) Theoretical studies on the necessary number of components in mixtures. 4. Number of components and juvenile–mature correlations. Theor Appl Genet 73:53–60
Isik F, Li B, Frampton J (2003) Estimates of additive, dominance and epistatic genetic variances from a clonally replicated test of loblolly pine. For Sci 49:77–88
Isik F, Li B, Frampton J, Goldfarb B (2004) Efficiency of seedlings and rooted cuttings for testing and selection in Pinus taeda. For Sci 50:44–53
Isik F, Goldfarb B, Lebude A, Li B, McKeand S (2005) Predicted genetic gains and testing efficiency from two loblolly pine clonal trials. Can J For Res 35:1754–1766
King JN, Carson MJ, Johnson GR (1998) Analysis of disconnected diallel mating designs. II—results from a third generation progeny test of the New Zealand radiata pine improvement programme. Silvae Genet 47:80–87
Kumar S (2006) Correlation between clonal means and open-pollinated seedling progeny means and its implications for radiata pine breeding strategy. Can J For Res 36:1968–1975
Libby WJ (1969) Clonal selection and an alternative seed orchard scheme. Silvae Genet 13:32–40
Libby WJ, Jund E (1962) Variances associated with cloning. Heritability 17:533–540
Libby WJ, Rauter RM (1984) Advantages of clonal forestry. The For Chron 6:145–149
Lu PX, White TL, Huber DA (1999) Estimating type B genetic correlations with unbalanced data and heterogeneous variances for half-sib experiments. For Sci 45:562–572
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer, Sunderland, p 980
Magnussen S (1993) Growth differentiation in white spruce crop tree progenies. Silvae Genet 42:258–266
Merrill RE, Mohn CA (1985) Heritability and genetic correlations for stem diameter and branch characteristics in white spruce. Can J For Res 15:494–497
Mullin TJ (1990) Genetic parameters for clonal selection of black spruce and implications for breeding. Ph.D. dissertation. University of New Brunswick, Fredericton, NB
Mullin TJ, Park YS (1992) Estimating genetic gains from alternative breeding strategies for clonal forestry. Can J For Res 22:14–23
Mullin TJ, Park YS (1994) Genetic parameters and age–age correlations in a clonally replicated test of black spruce after 10 years. Can J For Res 24:2330–2341
Mullin TJ, Adams GW, Simpson JD, Tosh KJ, Greenwood MS (1995) Genetic parameters and correlation in tests of open-pollinated black spruce families in field and retrospective nursery test environments. Can J For Res 25:270–285
NBTIC (1999) Black spruce clonal tests. Establishment Report No. 57. Fredericton, NB
Nienstaedt H (1984) Inheritance and correlations of frost injury, growth, flowering, and cone characteristics in white spruce, Picea glauca (Moench) Voss. Can J For Res 15:498–504
Nienstaedt H, Riemenschneider DE (1985) Changes in heritability estimates with age and site in white spruce, Picea glauca (Moench) Voss. Silvae Genet 34:34–41
Park YS, Bonga JM, Mullin TJ (1998) Clone forestry. In: Mandal AK, Gibson GL (eds) Forest genetics and tree improvement. CBS Publishers & Distributors, New Delhi, pp 143–167
Paul AD, Fistyer GS, Caldwell T, McRae J (1997) Trends in genetic and environmental parameters for height, diameter, and volume in a multilocation clonal study with loblolly pine. For Sci 43:87–98
Rouland H, Wellendore H, Werner M (1986) A selection experiment for height growth with cuttings of Picea abies (L.) Karst. Scand J For Res 1:293–302
Stonecypher RW, McCullough RB (1986) Estimates of additive and non-additive genetic variances from a clonal diallel of Douglas-fir Pseudotsuga menziesii [Mirb] Franco. In: Conference proceedings IUFRO joint meeting of working parties on breeding theory, progeny testing and seed orchards, Oct. 13–17, 1986. Williamsburg, VA, pp 211–227
Sulzer AM, Greenwood MS, Livingston WH, Adams G (1993) Early selection of black spruce using physiological and morphological criteria. Can J For Res 23:657–664
Weng YH, Tosh KJ, Park YS, Fullarton MS (2007) Application of nursery testing in long-term white spruce improvement programs. North J Appl For 24:296–300
Wilcox JR, Farmer RE (1968) Heritability and C effects in early growth of eastern cottonwood cuttings. Heritability 23:239–245
Williams DJ, Dancik BP, Pharis RP (1987) Early progeny testing and evaluation of controlled crosses of black spruce. Can J For Res 17:1442–1450
Wricke G, Weber WE (1986) Quantitative genetics and selection in plant breeding. Walter de Gruyter, Berlin, p 406
Wu HX (1998) Study of early selection in tree breeding. I. Advantage of early selection through increase of selection intensity and reduction of field test size. Silvae Genet 47:146–155
Xiang B, Li BL, McKeand S (2003) Genetic gain and selection efficiency of loblolly pine in three geographic regions. For Sci 49:196–208
Yamada Y (1962) Genotype by environment interaction and genetic correlation of the same trait under different environments. Jpn J Genet 37:498–509
Zobel BJ, Talbert JT (1984) Applied forest tree improvement. John Wiley & Sons, Inc. New York, USA
Acknowledgments
We would like to thank Dale Simpson and John Major for valuable comments on an earlier version of the manuscript and Caroline Simpson for editing. We would like to express our appreciation to Ian MacEacheron, CFS, and New Brunswick DNR tree improvement technicians Craig Carr, Mark Mazerolle, Kirk Beers, Roberto Florean, and Dan Phillips for the help in data collection. Thanks also went to Dr. Luis Apioloza for his help in running ASREML program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Aitken
Rights and permissions
About this article
Cite this article
Weng, Y.H., Park, Y.S., Krasowski, M.J. et al. Partitioning of genetic variance and selection efficiency for alternative vegetative deployment strategies for white spruce in Eastern Canada. Tree Genetics & Genomes 4, 809–819 (2008). https://doi.org/10.1007/s11295-008-0154-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-008-0154-0