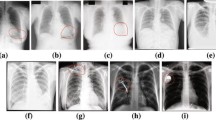
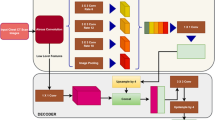
Abstract
Segmentation of lung fields is an important pre-requisite step in chest radiographic computer-aided diagnosis systems as it precisely defines the region-of-interest on which different operations are applied. However, it is immensely challenging due to extreme variations in shape and size of lungs. Manual segmentation is also prone to large inter-observer and intra-observer variations. Thus, an automated method for lung field segmentation with sufficiently high accuracy is unsparingly required. This paper presents a deep learning-based fully convolutional encoder-decoder network for segmenting lung fields from chest radiographs. The major contribution of this work is in the unique design of the encoder-decoder network that makes it especially suitable for lung field segmentation. The proposed network is trained, tested and evaluated on publicly available standard datasets. The result of evaluation indicates that the performance of the proposed method, i.e. accuracy of 98.73% and overlap of 95.10%, is better than state-of-the-art methods.










Similar content being viewed by others
References
Alexander Kalinovsky, A., & Kovalev, V. (2016). Lung image segmentation using deep learning methods and convolutional neural networks. In XIII International Conference on Pattern Recognition and Information Processing, Minsk: Publishing Center of BSU.
Annangi, P., Thiruvenkadam, S., Raja, A., Xu, H., Sun, X., & Mao, L. (2010). A region based active contour method for x-ray lung segmentation using prior shape and low level features. In 2010 IEEE international symposium on biomedical imaging: from nano to macro, pp. 892–895.
Arbabshirani, M. R., Dallal, A. H., Agarwal, C., Patel, A., & Moore, G. (2017). Accurate segmentation of lung fields on chest radiographs using deep convolutional networks. In SPIE medical imaging (pp. 1013,305–1013,305). International Society for Optics and Photonics.
Armato, S. G., Giger, M. L., & MacMahon, H. (1998). Automated lung segmentation in digitized posteroanterior chest radiographs. Academic Radiology, 5(4), 245–255.
Badrinarayanan, V., Handa, A., & Cipolla, R. (2015). Segnet: A deep convolutional encoder-decoder architecture for robust semantic pixel-wise labelling. arXiv preprint arXiv:1505.07293
Badrinarayanan, V., Kendall, A., & Cipolla, R. (2015). Segnet: A deep convolutional encoder-decoder architecture for image segmentation. arXiv preprint arXiv:1511.00561
Berbaum, K. S., Krupinski, E. A., Schartz, K. M., Caldwell, R. T., Madsen, M. T., Hur, S., et al. (2015). Satisfaction of search in chest radiography 2015. Academic Radiology, 22(11), 1457–1465.
Breuninger, M., van Ginneken, B., Philipsen, R. H., Mhimbira, F., Hella, J. J., Lwilla, F., et al. (2014). Diagnostic accuracy of computer-aided detection of pulmonary tuberculosis in chest radiographs: A validation study from sub-Saharan Africa. PLoS ONE, 9(9), e106,381.
Candemir, S., Jaeger, S., Palaniappan, K., Antani, S., & Thoma, G. (2012). Graph cut based automatic lung boundary detection in chest radiographs pp. 7–9.
Candemir, S., Jaeger, S., Palaniappan, K., Musco, J. P., Singh, R. K., Xue, Z., et al. (2014). Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Transactions on Medical Imaging, 33(2), 577–590. https://doi.org/10.1109/TMI.2013.2290491.
Chabi, M. L., Borget, I., Ardiles, R., Aboud, G., Boussouar, S., Vilar, V., et al. (2012). Evaluation of the accuracy of a computer-aided diagnosis (CAD) system in breast ultrasound according to the radiologist’s experience. Academic Radiology, 19(3), 311–319.
Elemraid, M. A., Muller, M., Spencer, D. A., Rushton, S. P., Gorton, R., Thomas, M. F., et al. (2014). Accuracy of the interpretation of chest radiographs for the diagnosis of paediatric pneumonia. PLoS ONE, 9(8), e106,051.
van Ginneken, B., Stegmann, M. B., & Loog, M. (2006). Segmentation of anatomical structures in chest radiographs using supervised methods: A comparative study on a public database. Medical Image Analysis, 10(1), 19–40.
He, K., Zhang, X., Ren, S., & Sun, J. (2015). Deep residual learning for image recognition. arXiv preprint arXiv:1512.03385
Ioffe, S., & Szegedy, C. (2015). Batch normalization: Accelerating deep network training by reducing internal covariate shift. CoRR abs/1502.03167. arXiv:1502.03167
Jaeger, S., Candemir, S., Antani, S., Wáng, Y. X. J., Lu, P. X., & Thoma, G. (2014). Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quantitative Imaging in Medicine and Surgery, 4(6), 475–477.
Kim, J., Lee, S., Lee, G., Park, Y., & Hong, Y. (2016). Using a method based on a modified k-means clustering and mean shift segmentation to reduce file sizes and detect brain tumors from magnetic resonance (mri) images. Wireless Personal Communications, 89(3), 993–1008.
Kingma, D., & Ba, J. (2015). Adam: A method for stochastic optimization. In Proceedings of the 3rd international conference on learning representations (ICLR).
Kok, E. M., Abed, A., & Robben, S. G. F. (2017). Does the use of a checklist help medical students in the detection of abnormalities on a chest radiograph? Journal of Digital Imaging, 30(6), 726–731. https://doi.org/10.1007/s10278-017-9979-0.
Kosack, C., Spijker, S., Halton, J., Bonnet, M., Nicholas, S., Chetcuti, K., Mesic, A., Brant, W., Joekes, E., & Andronikou, S. (2017). Evaluation of a chest radiograph reading and recording system for tuberculosis in a hiv-positive cohort. Clinical Radiology 72(6), 519.e1–19.e9. https://doi.org/10.1016/j.crad.2017.01.008. http://www.sciencedirect.com/science/article/pii/S0009926017300363
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems, pp. 1097–1105.
Li, L., Zheng, Y., Kallergi, M., & Clark, R. A. (2001). Improved method for automatic identification of lung regions on chest radiographs. Academic Radiology, 8(7), 629–638.
McNitt-Gray, M. F., Huang, H., & Sayre, J. W. (1995). Feature selection in the pattern classification problem of digital chest radiograph segmentation. IEEE Transactions on Medical Imaging, 14(3), 537–547.
Melendez, J., Sánchez, C. I., Philipsen, R. H., Maduskar, P., Dawson, R., Theron, G., et al. (2016). An automated tuberculosis screening strategy combining X-ray-based computer-aided detection and clinical information. Scientific Reports, 6, 25,265.
Mittal, A., Hooda, R., & Sofat, S. (2017). Lung field segmentation in chest radiographs: A historical review, current status, and expectations from deep learning. IET Image Processing, 11(11), 937–952. https://doi.org/10.1049/iet-ipr.2016.0526.
Myles-Worsley, M., Johnston, W. A., & Simons, M. A. (1988). The influence of expertise on X-ray image processing. Journal of Experimental Psychology. Learning, Memory, and Cognition, 14(3), 553–557.
Novikov, A. A., Major, D., Lenis, D., Hladuvka, J., Wimmer, M., & Buhler, K. (2017). Fully convolutional architectures for multi-class segmentation in chest radiographs. arXiv preprint arXiv:1701.08816
Oliveira, L. L., Silva, S. A., Ribeiro, L. H., de Oliveira, R. M., Coelho, C. J., & S Andrade, A. L. (2008). Computer-aided diagnosis in chest radiography for detection of childhood pneumonia. International Journal of Medical Informatics, 77(8), 555–564.
Plankis, T., Juozapavicius, A., Stašiene, E., & Usonis, V. (2017). Computer-aided detection of interstitial lung diseases: A texture approach. Nonlinear Analysis, 22(3), 404–411.
Rahman, M. T., Codlin, A. J., Rahman, M. M., Nahar, A., Reja, M., Islam, T., Qin, Z. Z., Khan, M. A. S., Banu, S., & Creswell, J. (2017). An evaluation of automated chest radiography reading software for tuberculosis screening among public- and private-sector patients. European Respiratory Journal, 49(5), 1602159.
Robinson, J. W., Brennan, P. C., Mello-Thoms, C., & Lewis, S. J. (2016). Reporting instructions significantly impact false positive rates when reading chest radiographs. European Radiology, 26(10), 3654–3659.
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-net: Convolutional networks for biomedical image segmentation. In International conference on medical image computing and computer-assisted intervention (pp. 234–241). New York: Springer.
Shaw, N., Hendry, M., & Eden, O. (1990). Inter-observer variation in interpretation of chest X-rays. Scottish Medical Journal, 35(5), 140–141.
Shi, Y., Qi, F., Xue, Z., Chen, L., Ito, K., Matsuo, H., et al. (2008). Segmenting lung fields in serial chest radiographs using both population-based and patient-specific shape statistics. IEEE Transactions on Medical Imaging, 27(4), 481–494.
Shi, Z., Zhou, P., He, L., Nakamura, T., Yao, Q., & Itoh, H. (2009). Lung segmentation in chest radiographs by means of gaussian kernel-based fcm with spatial constraints. In Sixth international conference on fuzzy systems and knowledge discovery, 2009. FSKD’09 (Vol. 3, pp. 428–432).
Shiraishi, J., Katsuragawa, S., Ikezoe, J., Matsumoto, T., Kobayashi, T., Komatsu, Ki, et al. (2000). Development of a digital image database for chest radiographs with and without a lung nodule: Receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. American Journal of Roentgenology, 174(1), 71–74.
Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. In Proceedings of international conference on learning representations. arXiv:1409.1556
Sivaganesan, D. (2017). Wireless distributive personal communication for early detection of collateral cancer using optimized machine learning methodology. Wireless Personal Communications, 94(4), 2291–2302.
Snchez Morillo, D., Len Jimnez, A., & Moreno, S. A. (2013). Computer-aided diagnosis of pneumonia in patients with chronic obstructive pulmonary disease. Journal of the American Medical Informatics Association, 20(e1), e111–e117. https://doi.org/10.1136/amiajnl-2012-001171.
Suzuki, K. (2017). Computer-aided detection of lung cancer. In Image-based computer-assisted radiation therapy (pp. 9–40). New York: Springer.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V., & Rabinovich, A. (2015). Going deeper with convolutions. In Computer vision and pattern recognition (CVPR). arXiv:1409.4842
Tarrac, S. E. (2009). A systematic approach to chest x-ray interpretation in the perianesthesia unit. Journal of PeriAnesthesia Nursing, 24(1), 41–49. https://doi.org/10.1016/j.jopan.2008.11.001. http://www.sciencedirect.com/science/article/pii/S1089947208003298.
Toriwaki, J. I., Suenaga, Y., Negoro, T., & Fukumura, T. (1973). Pattern recognition of chest X-ray images. Computer Graphics and Image Processing, 2(3), 252–271. https://doi.org/10.1016/0146-664X(73)90005-1. http://www.sciencedirect.com/science/article/pii/0146664X73900051.
Tsujii, O., Freedman, M. T., & Mun, S. K. (1998). Automated segmentation of anatomic regions in chest radiographs using an adaptive-sized hybrid neural network. Medical Physics, 25(6), 998–1007.
Van Ginneken, B., Frangi, A. F., Staal, J. J., ter Haar Romeny, B. M., & Viergever, M. A. (2002). Active shape model segmentation with optimal features. IEEE Transactions on Medical Imaging, 21(8), 924–933.
Van Ginneken, B., Stegmann, M. B., & Loog, M. (2006). Segmentation of anatomical structures in chest radiographs using supervised methods: A comparative study on a public database. Medical Image Analysis, 10(1), 19–40.
W.S.H.M, Wan Ahmad, Zaki, W. M. D. W., & Ahmad Fauzi, M. F. (2015). Lung segmentation on standard and mobile chest radiographs using oriented Gaussian derivatives filter. Biomedical Engineering Online, 14, 20. https://doi.org/10.1186/s12938-015-0014-8.
Wang, J., & Perez, L. (2017). The effectiveness of data augmentation in image classification using deep learning. Tech. rep., Technical report.
Xu, T., Mandal, M., Long, R., & Basu, A. (2009). Gradient vector flow based active shape model for lung field segmentation in chest radiographs. In Proceedings of annual international conference of the IEEE engineering in medicine and biology society. IEEE engineering in medicine and biology society (Vol. 2009, p. 3561).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mittal, A., Hooda, R. & Sofat, S. LF-SegNet: A Fully Convolutional Encoder–Decoder Network for Segmenting Lung Fields from Chest Radiographs. Wireless Pers Commun 101, 511–529 (2018). https://doi.org/10.1007/s11277-018-5702-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-018-5702-9