Abstract
Lung segmentation algorithms play a significant role in segmenting theinfected regions in the lungs. This work aims to develop a computationally efficient and robust deep learning model for lung segmentation using chest computed tomography (CT) images with DeepLabV3 + networks for two-class (background and lung field) and four-class (ground-glass opacities, background, consolidation, and lung field). In this work, we investigate the performance of the DeepLabV3 + network with five pretrained networks: Xception, ResNet-18, Inception-ResNet-v2, MobileNet-v2 and ResNet-50. A publicly available database for COVID-19 that contains 750 chest CT images and corresponding pixel-labeled images are used to develop the deep learning model. The segmentation performance has been assessed using five performance measures: Intersection of Union (IoU), Weighted IoU, Balance F1 score, pixel accu-racy, and global accuracy. The experimental results of this work confirm that the DeepLabV3 + network with ResNet-18 and a batch size of 8 have a higher performance for two-class segmentation. DeepLabV3 + network coupled with ResNet-50 and a batch size of 16 yielded better results for four-class segmentation compared to other pretrained networks. Besides, the ResNet with a fewer number of layers is highly adequate for developing a more robust lung segmentation network with lesser computational complexity compared to the conventional DeepLabV3 + network with Xception. This present work proposes a unified DeepLabV3 + network to delineate the two and four different regions automatically using CT images for CoVID-19 patients. Our developed automated segmented model can be further developed to be used as a clinical diagnosis system for CoVID-19 as well as assist clinicians in providing an accurate second opinion CoVID-19 diagnosis.
Similar content being viewed by others
1 Introduction
Recent report from WHO reports that lung disorders are the third prominent cause of death and about three million people across the globe lose their lives each year due to different types of lung disorders [1]. Lung diseases are mostly caused due to infections by either bacteria or viruses. Also smoking, air pollution and genetic factors affect the lung. If the lungs that have been negatively affected by a bacterial or viral infection are not treated within a short time frame, the lungs can suffer severe consequences. More recently, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has been shown to be an emerging human infectious disease that can have detrimental consequences on the lungs and has a significant infectious rate. Early diagnosis of lung infections through advanced clinical diagnosis systems using artificial intelligence (AI) methods could save people before more damage to the lungs have can occur. In recent years, due to the lack of medical specialists, there is a huge demand for developing intelligent computerized clinical diagnosis systems (CCDS) using AI methods to assist physicians in an accurate clinical diagnosis and treatment. Medical imaging tools (i.e., chest x-ray (CXR), computerized tomography (CT), and magnetic resonance imaging (MRI)) play a vital role in CCDS for diagnosing lung diseases. They provide more valuable information about lung infection with higher precision and accuracy [2]. Such imaging modalities are very helpful for the diagnosis of lung diseases. Specifically, CT images give clearer and more detailed information about the lungs than CXR [3]. To develop CCDS, it is essential to segment the lungs from medical images such as CXR or CT scan for better clinical interpretation. Hence, the segmentation process is important, and it is used for diagnosing different types of pathologies and severity levels of lung diseases. In the segmentation process, the researchers are interested in investigating the infected regions in the lungs using the region of interest (ROI). There are several lung segmentation algorithms proposed using conventional approaches, namely gray-level co-occurrence matrix (GLCM), watershed method, thresholding method, edge-based method, local binary patterns (LBP), region-based method, and others have been proposed in the literature [4, 5]. In the thresholding method, different types of thresholding algorithms are used for segmentation such as global thresholding, Otsu’s binarization, adaptive thresholding [6]. These methods have several limitations and one of the most evident limitations is that some of the most vital information in the images related to diagnosis can be eliminated due to improper selection of threshold values. Up to this work and for the most part, researchers have used either a biased or heuristic approach to select the threshold values [7]. Region-based methods like region growing, region merging and splitting, etc. are computationally inefficient and are highly sensitive to noise [8]. The region-based methods and edge-based methods are not successful in segmenting the lungs using noisy images. Clustering-based techniques such as fuzzy C-means and K-means are unable to find an optimal value of cost function to perform efficient lung segmentation. Also, in water-shed methods, it is complex to calculate the gradient for ridges. Some of the major limitations in the conventional segmentation algorithms are: (a) Highly challenging to select the exceptional thresholding value (or) hyperparameters, (b) Different assumptions and approximations are used in threshold parameter selection, (c) Lack of expertise and intelligence in understanding the image descriptions, and (d) Segmentation for the most part is accomplished using a preferred number of features. In some cases, the process of segmentation also segments the unwanted information and highly sensitive to noise. Statistical methods and descriptors are mostly utilized by the researchers to wrest features from the lung regions in MRI and CT images [9]. Consequently, the above-mentioned approaches require more processing time, and can lead to, many times, extraction of irrelevant and redundant information.
Recently, deep neural networks attracted many researchers and they proposed different types of deep learning algorithms for semantic lung segmentation [10, 11]. To achieve better segmentation, deep neural networks with encoder and decoder overcome the overlapping issue of the model gets trainedwith both forward and backward propagation and hence checks twice to reduce the loss value. Some of the major advantages of DNN are [5]: (i) fewer numbers of required images can be sufficient to design a network, as well as provide better performance than many conventional methods. (ii) There is no need to feed all points and edge features for segmentation. (iii) Convolutional nets can extract deep features to effectively segment the images. (iv) The input of the DNN architecture looks like a contracting path to extract high-level information, and the output has a symmetric expanding path to precisely localize the information, and (v) annotated lesions can be extracted without handcrafted features and labeling. Unlike conventional methods, the DNN can be used todirectly segment the lungs using either CXR or CT images by extracting deep features without prior knowledge requirement.
In the literature, lung segmentation using medical images have been mostly performed using Convolutional neural networks (CNNs) than other DNNs [12, 13]. In general, segmentation can be performed either semantically or instance-based. In instance segmentation, multiple objects of the same class are treated as distinct individual instances, which is unlike objects of the same class which are considered as the same entity in semantic segmentation [14]. In clinical diagnosis, semantic segmentation is widely used. Some of the most used segmentation networks besides CNN are, DeepLabv3, U-Net, SegNet, andFully Convolutional Network (FCN) [9, 13, 15]. DeepLabv3 was developed by Google, it gives exceptional performance in semantic segmentation compared to other segmentation networks and it assigns semantic labels to each pixel inthe image [16]. Recently, DeepLabV3 performed better than other DNNs for segmentation using MRI images [17]. The DeepLabV3 architecture consists of an atrous encoder and can encode multi-scale contextual information. As well as having the ability to decode for concatenation of lower-level features. Their four versions of DeepLab network have been developed by Google, DeepLabv1, DeepLabv2, DeepLabv3, and DeepLabv3+ [18]. DeepLabv3+ is one of the faster and stronger DNN’s compared to its earlier version. The major differences between different DeepLab networks are given in Table 1.
Though the performance of the DeepLabV3+ network is highly competitive than other DNNs and also with the other versions of DeepLab, the network has Xception network as a pretrained network with nearly 23 million parameters and has 71 layers. Hence, the computational efficiency of the model is lower compared to other DNN’s. Hence, it is highly essential to investigate theDeepLabV3+ model with different pretrained networks to improve its computational efficiency without affecting its performance. For the most part, earlier work focused on segmenting the lung using CT images without considering different sub-regions of lungs during segmentation. The changes in lung segmentation network architecture do not improve the quality of segmentation and occasionally give the bad performance [19]. Most of the semantic lung segmentation algorithms proposed in the literature are focused on segmenting lungs from the background using CXR and chest CT-scan images for diagnosis. A very limited number of the literature has focused on multi-class segmentationusing medical images. To develop an intelligent clinical diagnosis system, it is highly essential to segment different regions of the lungs to identify the level of infection/severity through automated methods [20]. This work aims to design and develop a lightweight and computationally efficient DNN architecture for multi-class lung segmentation using chest CT images. The DeepLabv3+ network is used with four pretrained networks (ResNet, Xception, MobileNet-v2, and Inception-ResNet) to compare its performance in lung segmentation using chest CT images. The input images fed to the network are segmented into two class (lung and background) and four-class (lung, ground-glass opacities, background, and consolidation). The performance of the network is also tested with two different image sizes, namely 256 × 256 and 512 × 512 to compare the performance of the networks for lung segmentation. The deep neural network hyperparameters are selected basedon empirical analysis and finally, the DeepLabV3+ with the best-pretrained network has been reported in this work for two-class and four-class automated lung segmentation tasks.
The major contributions of this present work are given as follows:
-
1.
Investigated the DeepLabV3+ DNN with different transfer learningapproaches and proposed a DeepLabV3+ network with ResNet-18 that requires a lesser number of layers than conventional DeepLabV3+ net-work. The proposed network requires fewer network parameters and achieved higher accuracy in semantic segmentation compared to the originalDeepLabV3+ network.
-
2.
Two different image sizes of chest CT images with different transfer learning approaches are investigated and employed grid search method to identifythe optimal network parameters for DeepLabV3 + network.
-
3.
Proposed a unified segmentation model to delineate the two (lung andbackground) and four different regions (lung, background, ground-glassopacities, and consolidation) automatically using CT images for CoVID-19detection system.
The rest of the paper is organized as follows. Literature review of earlier works related to semantic lung segmentation from chest CT-images is presented in Sect. 2. Detailed methodology of the present work is provided in Sect. 3. The experimental results obtained, discussion, limitations, and future works are described in Sect. 4. Finally, the conclusion of the present work is given in Sect. 5.
2 Related work
This section reviews the related work of lung segmentation methods developed using conventional and deep learning methods. One work describes that Optimal thresholding methods based lung segmentation in cancer CT scan images for developing a computer-aided diagnosis (CAD) system. The value of the threshold for segmenting the lung region and background was computed using the mean pixel values in the specific region. After thresholding, the region growing, and connectivity analysis was performed for lung segmentation [21]. Gray-level thresholding was used to segment the lung region, a dynamic programming method is employed to select the right and left lungs, and finally, morphological operations are performed to smoothen the irregular boundaries around the lungs. In this work, they extracted the lung region using gray-level thresholding. In recent work, watershed thresholding based on markers approach was used for semantic lung segmentation in chest CT-scan images using DeepLabV3 network with MobileNetV2 as a backbone for binary classification (CoVID-19 /non-CoVID-19). A set of statistical features such as circulatory area and perimeter of the segmented area was computed from Gray Level Co-occurrence Matrix (GLCM) and the optimal features used for classification were selected through the use of a genetic algorithm (GA) [22]. Moreover, Wang et.al have designed and developed a fully convolutional network (FCN) based deep learning model for semantic segmentation. The proposed FCN achieved approximately 5% improvement compared to conven- tional U-Net for the same application [23]. The texture analysis method has been used for segmenting the lung field from chest CT images of patients with interstitial lung disease (ILD) [24]. The authors have proposed a thresholding method for lung segmentation and then the abnormal regions in the lung field have been identified through texture-feature images derived from the co-occurrence matrix. More recently, researchers have utilized the Gaussian mixture models (GMMs) to improve the graph cuts algorithm for lung field segmentation in chest CT images [19]. The expectation-maximization (EM) algorithm is used to compute the weight or probability of a pixel that belongs to the lungor background. Graphs created using those weights and minimum cut theory were used to perform the segmentation.
After the CoVID-19 pandemic, many researchers in the field focused effortson identifying the effect of CoVID-19 using medical images. The severity level of infection could be easily diagnosed if the system effectively segments different infected regions in the lungs such as consolidation, ground-glass opacities (GGO), multiple ground-glass opacity (mGGO), pleural effusion. In an article published recently, researchers used a multiple instance learning approach to identify three classes of x-ray images, namely, viral pneumonia, bacteria pneumonia, and healthy people [25]. In recent years, researchers have mostly preferred the deep learning algorithms due to the self-learning ability to learn the input image features and segment the infected lung regions as accurately as possible to develop an intelligent CAD. Consequently, a heat map has been generated by utilizing the average of the weights of the filters in the last convolutional layer to identify the most infected regions. The authors have reported a mean accuracy of 98% in detecting CoVID-19 and non-CoVID-19 classes. Recently, multi-level classification of 14 chest diseases including CoVID-19 using ResNet50 deep learning network is proposed [26]. Maximum accuracy of 92.52% for three classes (normal, CoVID-19, other chest diseases) and 66.34% for 14 classes is achieved.
Also more recently, a novel AI system has been developed for CoVID-19 detection using chest CT images and CNN [23]. Pu et.al have proposed to perform lung segmentation, disease detection, and progression analysis of CoVID-19 using serial chest CT-scan images [27]. The authors have used U-Net for automatic lung segmentation and elastic lung registration using a bidirectional elastic registration algorithm. Their proposed system achieved a maximum sensitivity of 95% and specificity of 84%. To overcome the limitation of handcrafted features-based lung segmentation, a Residual U-net with a false-positive removal algorithm is proposed in [28]. The algorithms have been developed by using the concept that a DNN with residual units extractsmore discriminative features compared to other lung segmentation techniques including conventional U-Net architecture. The use of residual block in U- Net reduced the issues related to performance degradation and different augmentation methods are used to improve the robustness of the network. An FCN with conditional random fields (CRF) is proposed for lung segmen- tation and the accuracy of segmentation is assessed through dice coefficient [29]. Deng et.al have proposed a novel lung infection segmentation network for CoVID-19 using CT images. The authors have proposed a fully supervised and semi-supervised network to segment the lung infected regions in CoVID- 19 CT images of labeled and unlabeled data [30]. In [31], the researchers have investigated four deep learning models such as DeepLabV3+, U-Net, Dilated Residual U-net, and Residual U-net, for automatic lung segmentation and U-Net gave an optimal performance compared to other models for segmentation by giving a dice score of 0.98%.
Munusamy et.al have proposed FractalCovNet architecture for segmentation of covid infected regions using chest CT-scan images and CXR [32]. The performance of the FractalCovNet has been compared with five different transfer learning approaches namely Segnet, U-net, ResNet-UNet, FCN, and DenseUNet. The CNN-LSTM network is used for CoVID-19 detection using chest CT images and classified the input CT images into three classes namely, CoVID-19 pneumonia, normal, and other pneumonia with a sensitivity of 94.9, 84, and 93.9%, respectively [33]. The Inception-V3 network is used to classify the input CXR and CT scan images into two (CoVID-19 pneumonia/non-CoVID-19) and four classes (viral pneumonia, bacteria pneumonia, CoVID-19, and normal) and achieved a maximum mean classification rate of 99.4 (two-class), and 98.1% (four class). Consequently, the authors have classified the CoVID-19 into three classes such as GGO, consolidation, and pleural effusion using DeepLabV3+ network with Xception-V2 as a backbone network [34]. Most of the works proposed have developed an efficient lung segmentation algorithm using deep neural networks and used different performance measures to assess the performance of segmentation. However, most of the earlier works focused on segmenting the lung region alone from the background CXR/CT scan images and a limited number of works performed multi-class segmentation. The detailed methodology of the proposed multi-class segmentation is given in Sect. 3.
3 Materials and methods
This section presents the detail about the database and methods employed to develop the automated lung segmentation using deep learning models.
3.1 Chest CT-scan database
The CoVID-19 chest CT images developed by China National Center forBio-information is used in this work for developing an automated lung segmentation algorithm [36]. It is one of the largest publicly available lung CTimage databases with three different classes namely; CoVID-19 (NCP), normal control (NC), and Common Pneumonia (CP). Database of CT images and metadata are constructed in different cohorts from the China Consortium of Chest CT Image Investigation (CC-CCII). It includes CT images of 929 NCP patients, 932 CP patients, and 818 NC subjects. Also, it contains the meta-data of this dataset which includes patient ID, sex, critical illness stage, scan ID, lung function, liver function, age, and time of progression.
3.2 DeepLabv3 + architecture
In this work, to develop automated lung detection, DeepLabv3+, which uses Atrous Spatial Pyramid Pooling (ASPP) module and encoder-decoder structure, is employed [18]. This type of convolution is proposed to eliminate the issues of the computational complexity of the deep neural network without affecting its performance. In DeepLabV3+, the performance of deep feature extraction has been improved by implementing the two-stage convolution called point-wise convolution and depth-wise convolution. This convolution method for a 2D image can be represented as Eq. 1 [18]
Here, i represents the pixel location in an image, the output of a convolution and convolution filter are represented as b, c(x), respectively. The r represents the atrous rate (stride) of convolution. In general, r = 1 is used in standard convolution, and further adjustments can be done to enlarge the field of view of filters. In this network, the ASPP module is used to extract rich contextual information through pooling operations and encoder-decoder structure is used to capture the sharper object boundaries by uniformly recovering the spatial information (Eq. 1). It offers an efficient mechanism to control the field-of-view and finds the best trade-off between accurate localization (small field-of-view) and context assimilation (large field- of-view). In this work, Eq. 1 has been implemented using different atrous rates (6, 12, and 18) with a constant convolution filter size of 3 × 3 (Fig. 1). A higher atrous rate allows the ASPP module to enlarge the field of view of filters to extract more information from the input image.
As encoder-for the task of image classification, the final feature map spatial resolution is usually 32 times smaller than the input image resolution. Hence, the output stride is maintained to 32 in the DeepLabV3+ architecture. The proposed architecture significantly improved the following factors: (i) feature extraction ability of the CNN, (ii) integration of contextual information, (iv) training procedures, and (iii) object scale modeling, [15]. DeepLab-V3+ is an extended version of DeepLab-V3 by adding a decoder module to refine the segmentation results, especially along object boundaries. Besides, it utilizes the depth-wise separable convolution to both ASPP [18] and decoder modules. Thereby, the DeepLabV3+ network is faster and has a stronger encoder- decoder network for automated semantic segmentation tasks compared toother deep neural networks.
The architecture of the DeepLabv3+ DNN use for automated lung segmentation is shown in Fig. 1. Several parallel atrous convolutions with different rates are applied to get multi-scale contextual information. The decoder module helps in the detailed object boundary recovery. In our work, the encoder features are bilinearly upsampled by a factor of 4 and concatenated with low-level features. After concatenation, 3×3 convolutions were applied to refine the features followed by another simple bilinear upsampling by a factor of 4.
3.3 Proposed methodology on lung segmentation
This section describes the design methodology of a novel DeepLabv3+ network using different pretrained networks for lung segmentation. Figure 2 shows theproposed methodology of lung segmentation using chest CT images (Table 1).
3.3.1 Database
In this work, we have utilized 750 images and corresponding pixel labeled images collected from 150 patients for developing the DNN based segmentation model, and the output images of the DNN are compared with pixel annotated images (masks) to estimate the performance of our proposed DNN in lung segmentation. A set of standard metrics are used to analyze the performance of the DNN. The details of the CT images database are presented in Table 2. All the images have different resolutions; varying from 512 × 512 to 2592 × 2592 and the images are saved in.jpg or.png format. In this work, we only considered the CT images and pixel-labeled images for automated lung segmentationusing the DeepLabV3 + network. The database contains many chest CT scan images from three different classes, but the radiologists only manually annotate750 images from 150 patients. Later, we grouped the pixel-labeled images and CoVID-19 images into two different groups such as two classes (background and lung), and four classes (background, lung, GGO, and consolidation). In the case of a two-class problem, pixel-labeled images are annotated as zero for background and one for lungs. Similarly, for four class segmentation, the pixels are annotated as zero for background, one for lung field, two for consolidation (CL), and three for ground-glass opacity (GGO) [35]. Hence, we utilized the 750 images and corresponding pixel-labeled images for this proposed segmentation model. Among them, 600 images (80%) (120 subjects) were chosen for training, 75 (10%) (15 subjects) images for validation, and 75 (10%) (15 subjects) images as test datasets. The ratio of data used for training, validation, and testing is 80:10:10. We have carefully grouped and verified that the training and testing images do not belong to the same subject to prevent bias.
3.3.2 Preprocessing
Furthermore, we have developed a deep neural network to perform the automated lung segmentation with two different resolutions such as 256 × 256 and 512 × 512. Pretrained models such as ResNet-18, ResNet-50, and MobileNet-v2 networks require the input image size to be 224 × 224. However, the input image size of 229 × 229 is required for Xception and Inception-ResNet-v2 networks. To maintain the consistency between the DeepLabV3+ with different pretrained networks, in this present work, we considered two image sizes such as 256 × 256 and 512 × 512. Hence, we performed an image resize operation in the preprocessing stage to generate the images with standard two dimensions (256 × 256 and 512 × 512) without affecting its texture information. To maintain uniformity, we have used the above two sizes for all the transfer learning approaches to evaluate its performance for automated lung segmentation. Similarly, we also performed the image resize operation on the pixel-labeled data for further analysis.
3.3.3 DeepLabV3 + network
The DeepLabV3+ DNN is used for lung segmentation in our work. Compared to other deep neural networks, DeepLabV3+ is the most efficient for segmentation and classification tasks. To investigate the performance of the DeepLabV3+ network in lung segmentation, we have used four different pretrained networks with a different number of layers such as ResNet [36], Inception [37], MobileNet-V2 [38], and Xception [39]. These pretrained networks are the best suitable for performing image segmentation and classification tasks using smaller datasets and have been trained with a huge number of images from the ImageNet database [39]. In our work, ResNet-18 with 18 layers, ResNet-50 with 50 layers, MobileNet-v2 with 53 layers, Xception with 71 layers, and Inception-ResNet-v2 with 164 layers are used with DeepLabV3+ for performance comparison of automated lung segmentation. Using the grid search method, the optimal hyperparameters are selected to obtain the best performance for lung segmentation. The detailed list of hyperparameters used is given in Sect. 4. The above process has been repeated for both two-class and four-class segmentation with two different image sizes to investigate the relationship between image size, network tuning parameters, semantic segmentation performance, etc.
3.3.4 Semantic segmentation performance measures
The following statistical measures are used to examine the performance of the DeepLabV3 + deep neural network in lung segmentation with different transferlearning approaches.
3.3.4.1 Accuracy
The accuracy measure is defined as a ratio of correctly classified pixels to the total number of pixels in that class, according to the ground truth. The pixel accuracy of each class can be computed by using Eq. 2. The average value of accuracy of all classes in all images is referred to as Mean Accuracy (ACC). Pixel accuracy (PA)/Global accuracy (GA) is the ratio of correctly classified pixels, regardless of class, to the total number of pixels and it can be computed using Eq. 3 [40, 41].
where TP-true positive, TN-true negative, FP-false positive, FN-false negative.
3.3.4.2 Boundary contour matching score
This score is mostly used to reflect the contour matching between the segmented images and the ground truth images. Boundary F1 (BF) score refers to the accuracy of contour matching between the predicted boundary of each class concerning its ground truth boundary (pixel-labeled image boundary) [11]. The average value of BF score of all classes in all images is referred as Mean BF Score (BFS). The numerical value of BF score in the range [0,1] and in the form of either scalar or vector. A score of 1 reflects the perfect match of the contours of the corresponding class in prediction with its ground truth contour. BF score can be calculated using the precision and recall values with a distance error tolerance [11]. The BFS can be computed by using the given Eq. 4 [11].
where
3.3.4.3 Intersection over union/Jaccard similarity coefficient
IoU is the ratio of correctly classified pixels to the total number of pixels in the ground truth and predicted pixels in that class and it can be computed using Eq. 7 [11]. For the given dataset, MeanIoU is the mean IoU score of all the images in all classes. Weighted-IoU shows the average IoU of each class, weighted by the number of pixels in that class.
4 Results and discussion
This section presents the experimental results of lung segmentation using our proposed DeepLabV3+ network with various transfer learning approaches. In this present work, we performed a semantic segmentation using a transfer learning approaches by utilizing the DeepLabv3+ model which is trained using various pretrained networks that are trained on the ImageNet database. The DeepLabV3+ network for lung segmentation has been trained using various transfer learning approaches (MobileNet-v2, ResNet-18, Xception, ResNet-50, and pretrained Inception-ResNet-v2) of two different image sizes (512 × 512 and 256 × 256). The Xception and pretrained Inception-ResNet-v2 models did not give a higher accuracy in semantic segmentation. Hence, the results of these networks are not reported in this paper. The accuracy of segmentation has been computed using five performance measures. The pixel counts used for training, validation, and testing are given in Table 3. The number of pixels in the background of CT images contributes 87%, and the lung field contributes 13% of total pixels for two classes. Besides the above-mentioned regions, the numberof pixels in GGO and consolidation contribute 1.11, and 0.75%, respectively, in the four-class segmentation. It is evident that the number of pixels used for training the network for different classes is highly unbalanced and there is no additional framework proposed in this present work for balancing the pixels in each class. The complete set of pixels in each class is fed to the network for training, validation, and testing, and the results obtained are given in this present section (Table 4).
In this work, we have trained the deep learning model with different ranges of hyperparameters. The optimal deep neural network hyperparameters were selected based on the grid-search method. Table 4 shows a set of hyperparameters selected for the fine-tuning, range of values used, and the optimal value of each hyperparameter that yielded the maximum segmentation accuracy. The values of the hyperparameter for each model are finalized after evaluating its performances in lung segmentation through performance measures. In this work, we used two optimizers, namely, ‘sgdm’ and ‘Adam’ in the output layer. However, the performance of the Stochastic Gradient Descent with Momentum(sgdm) optimizer did not perform satisfactorily as compared to the ‘Adam’ optimizer. Hence, we have only used the ‘Adam’ optimizer for all the networks. Also, various deep neural network models used gave the best performance for lung segmentation with an epoch value of 100 with a learning rate of 0.001. Besides, L2 regularization is applied to keep the weights and bias values smaller to prevent the models from over-fitting. The value of the regularization factor is assumed to be 0.0001 to reduce the network weights slowly.
We have trained the proposed model using 600 CT images and corresponding 600 pixel labeled images. The graphs of accuracy and loss versus a number of iterations obtained for our proposed DeepLabv3+ network during the training with ResNet18 and ResNet50 is shown in Fig. 3.
4.1 Two-class semantic segmentation using pretrained networks
Table 5 presents the summary of semantic segmentation performance measures obtained for two-class segmentation with an image size of 512 × 512. The ResNet-50 with a batch size of 8 gave a maximum value of Mean IoU, and Mean BF score of 0.9887 and 0.9901, respectively. Besides the MobileNet-v2, all other transfer learning approaches are limited to train the model with abatch size of either 8 or 16. This is mainly due to the limitations in the computation power of the computing system and input image size. Increasing the value of batch size above 16 in most of the pretrained networks causes mem- ory error during implementation. The maximum value of pixel accuracy after segmentation reached 0.9962 for ResNet18. The ResNet-50 network also gave a maximum global accuracy (0.9974) and weighted IoU (0.9950) compared to other pretrained networks. Though the ResNet50 network required more time compared to other networks, it gave the highest performance for two-class lung segmentation compared to other pretrained networks. The Xception and Inception-ResNet-V2 did not give a good performance for two-class segmentation with the image size of 512 × 512 and required more time (approximately 600 min) for training compared to other pretrained networks. Hence, the results obtained by the above two networks are not included in Tables 5 and 6. The best-performing performing model for an image size of 512 × 512, and 256 × 256 is highlighted by BOLD in Tables 5 and 6, respectively. The summary of the best-performing model and the segmentation performance measure of two different image sizes are given in Tables 8 and 9, respectively.
Table 6 shows the summary of semantic segmentation performance measures obtained for two-class segmentation with an image size of 256 × 256. In the case of smaller image size (256 × 256), the ResNet-18 pretrained network with a batch size of 16 gave an optimal performance in semantic lung segmentation compared to other pretrained networks. The experimental results also indicate that the ResNet18 with a batch size of 32 required less training time compared to other pretrained networks. Hence, it is highly evident that the increase in batch size reduces the training time of the deep neural network, and it is possible by using smaller size images during training. The maximum value of mean IoU is 0.9876, and the mean BF score is 0.99 for ResNet-18. Besides, the same network gave the highest global accuracy of 0.9972 and weighted IoU value of 0.9944 compared to other pretrained networks. During validation, the maximum accuracy of 99.71% is achieved using MobileNet-v2. Comparing the experimental results of two class semantic segmentation with two different image sizes, the values of the performance measures such as mean accuracy (pixel accuracy), mean IoU, mean BF score, and global accuracy obtained for smaller size images (256 × 256) are approximately closer to the values of largersize images (512 × 512). Besides, the training time required for smaller size images in ResNet-18 is much lesser than larger size images using the ResNet-50 network. Hence, the average performance of semantic segmentation fortwo-class with smaller size images is superior to larger size images. Table 7 presents the summary of semantic segmentation performance obtained for individual classes during two-class segmentation. ResNet-50 net- work efficiently segmented the lung field and background from the input chest CT images compared to other pretrained networks using an image size of 512 × 512. The ResNet50 network with a batch size of 8 gave the highest value of IoU for lung and background segmentation of 0.9803 and 0.9971, respectively compared to other networks. Similarly, the value of BF score for lung and back- ground field segmentation in chest CT image using ResNet-50 network is 0.9853 and 0.9950, respectively. However, the ResNet-18 network with a batch size of 16 gave better performance in lung field segmentation from a background using a smaller image size (256 × 256). The maximum mean IoU of 0.9784 and mean BF score of 0.9852 are achieved in lung segmentation. The highest value of pixel accuracy for background and lung field is achieved using Mobilenet- v2 network with a batch size of 16, and ResNet18 network with a batch size of 8. Besides, the ResNet18 network with a batch size of 16 gave a maximum value of Mean IoU (0.9968) for background segmentation. In conclusion, the ResNet-18 with a batch size of 16 performed well for two-class segmentation, and the higher resolution chest CT scan images gave a higher performance for lung segmentation compared to lower-resolution images. Specifically, there are no key differences between the performance measure values of higher and lower resolution images. Hence, the lower resolution images can be considered to evade the computational entanglement in image processing.
The summary of results obtained in Tables 5, 6, and 7 are given in 8 and Table 9, respectively. The segmentation performance measures of the best-performing deep learning model are highlighted by BOLD for an image size of 512 × 512 and 256 × 256 as reported in Table 7. Table 8 shows the details of the best-pretrained network, and its mean accuracy obtained for two-class segmentation of two different image sizes. Table 8 shows the details of the best-pretrained network and individual class accuracy obtained for two-class segmentation of two different image sizes. Using IoU and BF score results, ResNet-50 with a batch size of 8 gave an optimal performance for two-class segmentation with a larger size images compared to smaller-size images. The IoU and BF scores are considered as the two most influential performance measures in evaluating the accuracy of segmentation in medical images compared to other performance measures.The summary of the best-performing model and the segmentation performance measure of two different image sizes are given in Table 9.
4.2 Four-class semantic segmentation using pretrained networks
Table 10 presents the summary of semantic segmentation performance measures obtained for four classes using an image size of 256 × 256. The Mobilenet-V2 network with a batch size of 16 gave a maximum value of Mean IoU, Weighted Mean IoU, and Mean BF score of 0.7608, 0.9881, and 0.8985, respectively. The Mobilenet-v2 requires less training time (85 min 44 sec) compared to other pretrained networks, as well as obtaining a maximum mean global accuracy of 0.9935. The maximum batch size for ResNet is limited to 16, and only the MobileNet-v2 is used for training (with a batch size of 32). This is mainly due to the limitations in the computation power of the computing system used in this work, which is, in this case, dependent on the input image size. The Xception and Inception-ResNet-v2 did not yield good performance in four class segmentation with the image sizes of 512×512 and 256×256. It requires more time (approximately 660 min) for training compared to other pretrained networks. Hence, the results of the above two networks are not included in Tables 10 and 11. It is unlikely that the ResNet which performed superior for two-class segmentation did not give optimal performance for four-class segmentation. One of the major reasons for this may have to do with its ability to handle highly unbalanced datasets. The best-performing deep learning model for four class segmentation for two different image sizes (512 × 512, 256 × 256) and its performance measures are highlighted by BOLD in Tables 10 and 11.
Table 11 shows the summary of semantic segmentation performance measures obtained for four classes using an image size of 512 × 512. Among the pretrained networks, ResNet-50 with batch size 16 gave higher accuracy for four-class segmentation. The above network gave a maximum value of mean IoU, weighted mean IoU, and mean BF score of 0.7655, 0.9873, and 0.8985, respectively. The above network gave a maximum global accuracy of 0.9931 compared to other pretrained networks. The ResNet-18 with a batch size of 32 requires lesser training time compared to other pretrained networks. Hence, an increase in the batch size reduces the training time of the deep neural network, and it is possible by using smaller size images during training.
Table 12 shows the summary of semantic segmentation performance measures obtained for each class during the segmentation of four classes. The four classes presented in the chest CT images are lung field (LF), background (BG), ground-glass opacity (GGO), and consolidation (CL). The number of pixels in each class is different, the background has the largest number of pixels, and the consolidation section has the least number of pixels. For the image size 512 × 512, the ResNet-50 network with batch size 8 gave maximum mean pixel accuracy, background and GGO reported 0.9993, and 0.6811, respectively, among the other pretrained networks. The maximum mean pixel accuracy of 0.7802 is achieved for CL using ResNet-18 with batch size 16. Similarly, the ResNet-50 network with batch size 16 gave a maximum pixel accuracy of LFas 0.9908. The MobileNet-V2 with a batch size of 32 obtained the highest mean IoU for background and lung field as 0.9775, and 0.9559, respectively. MobileNet-V2 with a batch size of 16 gave the highest mean value of BF score for background and lung field. The ResNet-50 with a batch size of 8 obtained the highest mean BF score for GGO and ResNet-18 with a batch size of CL obtained the highest mean BF score of 0.7649 for consolidation. Finally, the ResNet-18 with a batch size of 16 gave an optimal performance in pixel accuracy, mean IoU, and mean BF score for segmenting consolidation region from CT images. However, the ResNet-50 with a batch size of 8 presented the optimal performance for segmenting background and GGO using chest CT images. In CoVID-19 detection, the segmentation of CL and GGO plays a significant role in detecting the severity of CoVID-19 infection. Hence, the ResNet pretrained networks outperformed other pretrained networks in yielding the highest performance measures in segmenting GGO and CL. It can be noted from IoU and BF score that, MobileNet-v2 pretrained networks are best suited for background and lung field segmentation.
Table 13 presents the summary of best performance obtained using the pretrained network for semantic segmentation of four classes. It can be noted from the experimental results that, MobileNet-v2 with a batch size 16 is best suited for large size images (512 × 512), and ResNet-50 with batch size 16 is bestsuited for lower resolution images (256 × 256). 14 shows the summary of the best individual class performance obtained using of best pretrained network for semantic segmentation for four classes. It can be noted that the background and lung fields have been accurately segmented with large-size images using MobileNet-v2 or ResNet pretrained networks. However, the GGO and consolidation are effectively segmented with small-size images using ResNetpretrained networks. For GGO and consolidation segmentation, the small size images obtained better performance than large size images. However, all the performance measures gave an accuracy approximately closer and there is no huge difference in the accuracy obtained among the pretrained networks. In the case of individual class accuracy, there is no consistency between the pretrained networks, and each network performed better on a specific class. The detailed results of the pretrained network which obtained the highest performance in each class segmentation for four classes are given in Table 14. Finally, the smaller size images gave better performance for GGO, and CL segmentation and larger size images presented a better performance for background and lung field segmentation. Though, there is no specific pretrained network that can be chosen for all tasks, the ResNet-50 with a batch size of 16 presented optimal performance for segmentation of four classes using chest CT images.
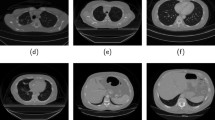
Tables 15 and 16 show the best results obtained for segmentation using DeepLabv3+ for the image size of 512 × 512, and 256 × 256 for two and four classes, respectively. As part of this study, the images in the dataset have already been annotated by expert radiologists [42]. We have used two different types of annotation in this study (i) two-class problem—the image pixels are labeled as zero for the background and one for the lungs (ii) four-class problem-the pixels are annotated as zero for the background, one for the lung field, two for consolidation (CL), and three for the GGO. As the proposed Deeplab V3+ network has been trained using pixel labels, it automatically annotates a test image based on the labels associated with the pixels. The original, resized, segmented, and ground truth images are shown. For two-class segmentation, the proposed DNN effectively segmented the lung field from the background. Visually, there is no major difference in the lung segmented images of 512 × 512, and 256 × 256 image sizes. Appendix (Tables 18 and 19) presents the semantic segmentation results of ten sample images which show four classes: lung field, GGO, consolidation, and background. The proposed DNN is able to effectively segment the GGO and consolidation region from CoVID-19 CT images. The performance comparison of the presented work with the state-of-the-art methods reported in the literature is presented in Table 17. In [35], the authors have used the same database as we used in this present work to perform lung segmentation using the DeepLabV3 network with ResNet as a backbone.ENet, U-net, and three-stage DNN framework have been used for lung segmentation in [35]. The U-net gave a better performance in lung segmentation ascompared to ENet [35] and the other three DNNs proposed by the researchers. They have achieved a maximum mean IoU of 0.7889, pixel accuracy of 0.9490 (background lung field), 0.5875 (GGO), and 0.6218 (consolidation). In their works, the DNNs have been trained and tested with an input image size of 512 × 512 and batch size of 4 to perform four-class segmentation (background, lung field, GGO, and consolidation). However, in [42], the authors have used more number of images and developed an AI-based CoVID-19 detection model (segmentation and classification). Five different types of DNNs such as U- net, DRUNET, FCN, SegNet, and DeepLabV3 have been used to segment the manually delineated CT images into seven regions (BG, LF, GGO, CL, pleural effusion, interstitial thickening, and pulmonary fibrosis). There are two performance metrics, namely, dice coefficient and pixel accuracy are computed to compare the performance of segmentation of different DNN’s. But, they have reported the performance of classification (two or three class) and not reported any performance metrics of segmentation. Finally, they concluded that the the DeepLabV3 model yielded better performance in lungs segmentation as compared to other DNN models.
In contrast with earlier works, we have employed the DeepLabV3+ networkusing different pretrained networks with different image sizes, as well as proposed a novel lightweight DNN model for performing two-class and four-class lung segmentation together. Compared to DeepLabv3, the DeepLabv3+ net-work decoder module is used to effectively obtain the segmentation results along the boundaries. The application of depth wise separable convolution in pooling and decoder module was used in order to obtain the segmentation results faster than conventional methods. Moreover, our network has been trained with the actual/original CT images instead of manually segmented images used in [39] to train the DNN. In our work, we have obtained maximum performance using DeepLab V3+ Network with ResNet-50 with a batch size of 8 for two class lung detection. Similarly, DeepLabV3+ Network with ResNet-50 and a batch size of 16 yielded optimum performance for four class segmentation tasks. Our work yielded a maximum mean accuracy of 0.9972, mean pixel accuracy of 0.9958, mean IoU of 0.9876, and mean BF score of 0.9900 for two-class segmentation using ResNet-50. The present work, also, reported highly competitive results for four-class segmentation using ResNet-50. The maximum mean global accuracy of 0.9931, mean accuracy of 0.8341, mean IoU of 0.7684, and mean BF score of 0.9008 is achieved using the ResNet-50. Hence, we have proposed a unified model for two class and four class segmentation tasks using DeepLab V3+ Network with ResNet-50 using a batch size of 8 (two classes) and 16 (four classes). To the bestof our knowledge, this is the first work to propose a model for two and four classes together.
In a recent study, Kaheel et al. used simple image processing (pre-processing, segmentation, feature extraction) and deep neural networks(ResNet50) to distinguish CoVID-19 from other viral pneumonia using chest CT scan images based on Corona score [43]. The author proposes simple machine learning algorithms (kNN, SVM) and deep neural networks (ResNet- 50, MobileNetv2) based on the detection of CoVID-19 using chest CT-scan images in [44]. In their study, the combination of DNN and SVM was found to be more accurate in detecting CoVID-19 than non-CoVID-19. Recently, Prakash et.al presented a method for CoVID-19 detection from chest X-rays based on super pixel-based segmentation and infection localization with SqueezeNet [13]. Researchers have used different types of deep learning algorithms to segment lungs from CT scan images, localize infected regions, and differentiate severity levels into four types, mild, moderate, severe, and critical [45]. Based on chest X-ray images and clinical data, one recent study differentiated CoVID-19 patients into low and high-risk groups using machine learning algorithms, resulting in a maximum mean F1 score of 92.88 [46].
However, the accuracy of our work is higher compared to all the earlier reported efforts with regards to lung segmentation. Also, we show how DeepLabV3 + network with ResNet pretrained network is highly efficient for two-class and four-class segmentation.
The proposed framework was implemented in a Desktop computer having Intel i7, an 8th generation processor with a speed of 3.20 GHz of 6 cores with 64 GB DDR4 RAM operating in the Windows operating system. The system is also equipped with four 8GB RTX2080 Graphical Processing Units (GPUs) for increased acceleration of the computation process.
4.3 Limitations and future works
Though the proposed deep neural network architecture yielded the highly accurate and promising lung segmentation performance, there are still some limitations to this work.
-
1.
Future work can compare other DNNs in lung image segmentation. In particular, lightweight DNN for CoVID-19 classification using chest CT-scanimages [13, 44].
-
2.
Future work can employ more CT scan images for training, validation and testing.
-
3.
Future work can compare performance using other databases, respectively. To overcome the above-mentioned limitations, in the future we intend to design a more robust DNN for lung segmentation with the following experiments.
-
1.
The performance of DeepLabV3+ network results will be compared withother networks such as capsule networks, CNN, SegNet, and U-net to gain7more insight into optimum efficiency, accuracy, and performance.
-
2.
Investigate the performance of the proposed network with multi-center/open-source lung CT image databases.
-
3.
Assess the performance of segmentation with other performance measuresfor better benchmarking.
-
1.
5 Conclusion
This work investigated the application of a DeepLabV3 + DNN with various pretrained networks for lung segmentation using CT images. The four most common types of pretrained networks such as ResNet-50, MobileNet-v2, ResNet-18, and Xception are used with 512 × 512 and 256 × 256 image sizes. These networks are used to perform two class (background and lung field) segmentation and four class (background, lung field, ground-glass opacities, consolidation) segmentation tasks. Our results confirm that there is no significant differences in lung segmentation performance between two different images sizes (256 × 256, 512 × 512) using the DeepLabV3 + network. We also show that the ResNet-50 with a batch size of 16 is best suited for four-class segmentation, and ResNet-18 with a batch size of 8 for two-class segmentation. The ResNet and MobileNet-v2 pretrained networks require less training time as compared to other pretrained networks such as Xception and Inception- ResNet-v2 networks. We proposed a novel unified segmentation model to delineate the two and four different regions automatically using CT images for COVID-19 detection. The proposed DeepLabV3 + network is developed for efficient lung segmentation using chest CT-scan images by selecting optimal pretrained network, image sizes, network hyperparameter selection, and computation time. The four-class segmentation of chest CT images can be used to develop an automated computerized clinical diagnosis system for CoVID- 19 to investigate the severity levels of CoVID-19 infections by delineating the affected lung region.
Data availability
The data that support the findings of this study are openly available in China National Center for Bio information at http://ncov-ai.big.ac.cn.
References
The global impact of respiratory disease – second edition. Forum of Inter-national Respiratory Societies,Sheffield, European Respiratory Society (2017)
Prayer F, R¨ohrich S, Pan J, Hofmanninger J, Langs G, Prosch H (2020) Artificial intelligence in lung imaging. Radiologe 60(1):42–47. https://doi.org/10.1007/s00117-019-00611-2
Benmalek E, Elmhamdi J, Jilbab A (2021) Comparing ct scan and chest x-ray imaging for covid-19 diagnosis. Biomed Eng Adv 1:100003. https://doi.org/10.1016/j.bea.2021.100003
Mansoor A, Bagci U, Foster B, Xu Z, Papadakis GZ, Folio LR, Udupa JK, Mollura DJ (2015) Segmentation and image analysis of abnormallungs at ct: Current approaches, challenges, and future trends. Radio-Graphics 35(4):1056–1076. https://doi.org/10.1148/rg.2015140232
Jangam E, Barreto AAD, Annavarapu CSR (2021) Automatic detectionof covid-19 from chest ct scan and chest x-rays images using deep learning, transfer learning and stacking. Appl Intell. https://doi.org/10.1007/s10489-021-02393-4
Essaf F, Li Y, Sakho S, Gadosey PK, Zhang T (2020) An improvedlung parenchyma segmentation using the maximum inter-class variancemethod (otsu), pp 204–212. https://doi.org/10.1145/3404555.3404647
Abd Elaziz M, Ewees AA, Oliva D (2020) Hyper-heuristic method for multi-level thresholding image segmentation. Expert Syst Appl 146:113201
Chandra SK, Bajpai MK (2019) Mesh free alternate directional implicitmethod based three dimensional super-diffusive model for benign braintumor segmentation. Comput Math Appl 77(12):3212–3223
Negi A, Raj ANJ, Nersisson R, Zhuang Z, Murugappan M (2020) Rda-unet-wgan: an accurate breast ultrasound lesion segmentation usingwasserstein generative adversarial networks. Arab J Sci Eng 45(8):6399–6410
Haque RI, Neubert J (2020) Deep learning approaches to biomedicalimage segmentation. Inform Med Unlock 18:100297. https://doi.org/10.1016/j.imu.2020.100297
Lateef F, Ruichek Y (2019) Survey on semantic segmentation using deeplearning techniques. Neurocomputing 338:321–348. https://doi.org/10.1016/j.neucom.2019.02.003
Vocaturo E, Zumpano E, Caroprese L (2021) Convolutional neural networktechniques on x-ray images for covid-19 classification. In: 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp 3113–3115. https://doi.org/10.1109/BIBM52615.2021.9669784
Prakash NB, Murugappan M, Hemalakshmi GR, Jayalakshmi M, Mahmud M (2021) Deep transfer learning for covid-19 detection and infectionlocalization with superpixel based segmentation. Sustain Cities Soc 75:103252. https://doi.org/10.1016/j.scs.2021.103252
Wang J, Xiong H, Wang H, Nian X (2020) Adscnet: asymmetric depthwiseseparable convolution for semantic segmentation in real-time. Appl Intell 50(4):1045–1056. https://doi.org/10.1007/s10489-019-01587-1
Yurtkulu SC, S¸ahin YH, Unal G (2019) Semantic segmentation with extended deeplabv3 architecture, pp 1–4. https://doi.org/10.1109/SIU.2019.8806244
Chen L-C, Papandreou G, Schroff F, Adam H (2017) Rethinking atrousconvolution for semantic image segmentation https://arxiv.org/abs/1706.05587 [cs.CV]
Furtado P (2021) Deep segmentation of abdominal organs from MRI: off-the-shelf architectures and improvements. Int Soc Opt Photon 11596:959–964
Chen L-C, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder-decoder with atrous separable convolution for semantic image segmen-tation. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y (eds) Computer Vision – ECCV 2018. Springer, Cham, pp 833–851
Dai S, Lu K, Dong J, Zhang Y, Chen Y (2015) A novel approach of lungsegmentation on chest ct images using graph cuts. Neurocomputing 168:799–807. https://doi.org/10.1016/j.neucom.2015.05.044
He K, Zhao W, Xie X, Ji W, Liu M, Tang Z, Shi Y, Shi F, Gao Y, Liu J, Zhang J, Shen D (2021) Synergistic learning of lung lobesegmentation and hierarchical multi-instance classification for automatedseverity assessment of covid-19 in ct images. Pattern Recogn 113:107828–107828. https://doi.org/10.1016/j.patcog.2021.107828
Javaid M, Javid M, Rehman MZU, Shah SIA (2016) A novel approachto cad system for the detection of lung nodules in ct images. Comput Methods Prog Biomed 135:125–139
Amin J, Sharif M, Anjum MA, Nam Y, Kadry S, Taniar D (2021) Diagnosis of covid-19 infection using three-dimensional semantic segmentationand classification of computed tomography images. Comput Mater Continua 68(2):2451–2467. https://doi.org/10.32604/cmc.2021.014199
Wang B, Jin S, Yan Q, Xu H, Luo C, Wei L, Zhao W, Hou X, Ma W, Xu Z et al (2021) Ai-assisted ct imaging analysis for covid-19 screen-ing: building and deploying a medical ai system. Appl Soft Comput 98:106897
Wang J, Li F (2009) Automated segmentation of lungs with severe inter-stitial lung disease in ct. Med Phys 36(10):4592–4599. https://doi.org/10.1118/1.3222872
Zumpano E, Fuduli A, Vocaturo E, Avolio M (2021) Viral pneumoniaimages classification by multiple instance learning: Preliminary results. In: Proceedings of the 25th International Database Engineering Applications Symposium. IDEAS ’21, pp 292–296. Association for ComputingMachinery, New York, NY, USA. https://doi.org/10.1145/3472163.3472170
Albahli SYG (2021) Fast and accurate detection of covid-19 along with 14 other chest pathologies using a multi-level classification: algorithm development and validation study. J Med Internet Res. https://doi.org/10.2196/23693
Pu J, Leader JK, Bandos A, Ke S, Wang J, Shi J, Du P, Guo Y, Wenzel SE, Fuhrman CR et al (2021) Automated quantification of covid- 19 severity and progression using chest ct images. Eur Radiol 31(1):436–446
Khanna A, Londhe ND, Gupta S, Semwal A (2020) A deep residual u-net convolutional neural network for automated lung segmentation in computed tomography images. Biocybern Biomed Eng 40(3):1314–1327
Alves JH, Neto PMM, Oliveira LF (2018) Extracting lungs from ct images using fully convolutional networks. In: 2018 International Joint Conference on Neural Networks (IJCNN), pp 1–8 (2018). IEEE
Fan D-P, Zhou T, Ji G-P, Zhou Y, Chen G, Fu H, Shen J, Shao L (2020) Inf-net: automatic covid-19 lung infection segmentation from ct images. IEEE Trans Med Imaging 39(8):2626–2637
Johannes H, Jeanny P, Sebastian R, Helmut P, Georg L (2020) Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp 4(1)
Munusamy H, Karthikeyan J, Shriram G, Revathi ST, Aravindku-mar S (2021) Fractalcovnet architecture for covid-19 chest x-ray image classification and ct-scan image segmentation. Biocybern Biomed Eng 41(3):1025–1038
Kara M, O¨ ztu¨rk Z, Akpek S, Turupcu A (2021) Covid-19 diagnosis from chest ct scans: a weakly supervised cnn-lstm approach. AI 2(3):330–341 https://doi.org/10.3390/ai2030020
El-bana A-K, Sharkas El-Bana S, Al-Kabbany A, Sharkas M (2020) A two- stage framework for automated malignant pulmonary nodule detection in CT scans. Diagnostics 10(3):131
Liu X, Wang K, Wang K, Chen T, Zhang K, Wang G (2020) Kiseg: a three-stage segmentation framework for multi-level acceleration of chest ct scans from covid-19 patients, pp 25–34. Springer
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 770–778
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C (2018) Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings ofthe IEEE Conference on Computer Vision and Pattern Recognition, pp 4510–4520
Chollet F (2017) Xception: deep learning with depthwise separable convolu-tions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 1251–1258
Studer L, Alberti M, Pondenkandath V, Goktepe P, Kolonko T, Fischer A, Liwicki M, Ingold R (2019) A comprehensive study of imagenetpre-training for historical document image analysis. In: 2019 International Conference on Document Analysis and Recognition (ICDAR), pp 720–725. IEEE
Sen S, Saha S, Chatterjee S, Mirjalili S, Sarkar, R (2021) A bi-stage featureselection approach for covid-19 prediction using chest ct images. Applied Intelligence. pp 1–16
Zhang K, Liu X, Shen J, Li Z, Sang Y, Wu X, Zha Y, Liang W, Wang C, Wang K et al (2020) Clinically applicable ai system for accu-rate diagnosis, quantitative measurements, and prognosis of covid-19pneumonia using computed tomography. Cell 181(6):1423–1433
Kaheel H, Hussein A, Chehab A (2021) Ai-based image processing for covid-19 detection in chest ct scan images. Front Commun Netw. https://doi.org/10.3389/frcmn.2021.645040
Canayaz M, Şehribanoğlu S, Özdağ R, Demir M (2022) COVID-19 diagnosis on CT images with Bayes optimization-based deep neural networks and machine learning algorithms. Neural Comput Appl 34(7):5349–5365. https://doi.org/10.1007/s00521-022-07052-4
Qiblawey Y, Tahir A, Chowdhury MEH, Khandakar A, Kiranyaz S, Rahman T, Ibtehaz N, Mahmud S, Maadeed SA, Musharavati F, Ayari MA (2021) Detection and severity classification of COVID-19 in CTimages using deep learning. Diagnostics (Basel) 11(5):893
Rahman T, Chowdhury MEH, Khandakar A, Mahbub ZB, Hos-sain MSA, Alhatou A, Abdalla E, Muthiyal S, Islam KF et al (2022) Biocxrnet: a robust multimodal stacking machine learning technique for mortality risk prediction of covid-19 patients using chest x-ray images and clinical data. https://doi.org/10.48550/ARXIV.2206.07595
Funding
This work is supported in part by the Kuwait Foundation for the Advancement of Sciences (KFAS), Kuwait through a priority research grant(Project Code: CN20-13QE-02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Murugappan, M., Bourisly, A.K., Prakash, N.B. et al. Automated semantic lung segmentation in chest CT images using deep neural network. Neural Comput & Applic 35, 15343–15364 (2023). https://doi.org/10.1007/s00521-023-08407-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-023-08407-1