Abstract
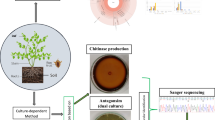
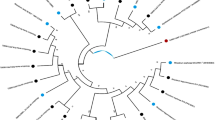
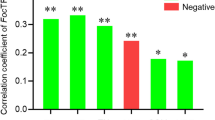
Endophytes have a wide range of potential in maintaining plant health and sustainable agricultural environmental conditions. In this study, we analysed the diversity of endophytic bacteria in four mulberry cultivars with different resistance capacity against bacterial wilt using metagenomic sequencing and culture-dependent approaches. We further assessed the role of 11 shared genera in the control of bacterial wilt of mulberry. The results of the present study showed that Actinobacteria, Firmicutes, and Proteobacteria were the three dominant phyla in all communities, with the representative genera Acinetobacter and Pseudomonas. The diversity analysis showed that the communities of the highly and moderately resistant varieties were more diverse compared to those of the weakly resistant and susceptible varieties. The control tests of mulberry bacterial wilt showed that Pantoea, Atlantibacter, Stenotrophomonas, and Acinetobacter were effective, with a control rate of over 80%. Microbacterium and Kosakonia were moderately effective, with a control rate between 50 and 80%. At the same time, Escherichia, Lysinibacillus, Pseudomonas, and Rhizobium were found to be less effective, with a control rate of less than 40%. In conclusion, this study provides a reasonable experimental reference data for the control of bacterial wilt of mulberry.







Similar content being viewed by others
Data availability
The metagenomic sequencing data of this study can be obtained at National Center for Biotechnology Information (NCBI) using accession number: PRJNA903404. The data of endophytic bacterial 16S rDNA isolated in this study have been submitted to NCBI with accession numbers: OP817191-OP817619.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Amorim AM, Nascimento JD (2017) Acinetobacter: an underrated foodborne pathogen? J Infect Dev Ctries 11(2):111–114
Bai J, Mo X, Zhou S, Lei F, Chen R, Gu W (1995) Guangxi mulberry pest and disease survey report. Guangxi Seric (02):24–27 (In China)
Barraquio WL, Revilla L, Ladha JK (1997) Isolation of endophytic diazotrophic bacteria from wetland rice. Plant Soi 194:15–24
Benhamou N, Kloepper JW, Quadt-Hallman A, Tuzun S (1996) Induction of defense-related ultrastructural modifications in Pea Root tissues inoculated with endophytic bacteria. Plant Physiol 112(3):919–929
Besaury L, Rémond C (2022) Culturable and metagenomic approaches of wheat bran and wheat straw phyllosphere is highlight new lignocellulolytic microorganisms. Lett Appl Microbiol 74(6):840–850
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Bredow C, Azevedo JL, Pamphile JA, Mangolin CA, Rhoden SA (2015) In silico analysis of the 16S rRNA gene of endophytic bacteria, isolated from the aerial parts and seeds of important agricultural crops. Genet Mol Res 14(3):9703–9721
Brown JK (2002) Yield penalties of disease resistance in crops. Curr Opin Plant Biol 5(4):339–344
Campista-León S, Cabanillas-Pacheco MJ, Delgado-Díaz LJ, Garcia-Guerrero JT, Peinado-Guevara LI (2022) Drug resistance and phylogenetic grouping of bacteria isolated from visitors’ hands in a secondary-care hospital. J Infect Dev Ctries 16(2):320–332
Conn VM, Walker AR, Franco CM (2008) Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol Plant Microbe Interact 21(2):208–218
Dai F, Wang Z, Luo G, Tang C (2016) Transcriptional analysis of different mulberry cultivars in response to Ralstonia solanacearum. Can J for Res 46(2):152–162
Dai F, Luo G, Li Z, Wei X, Wang Z, Lin S, Tang C (2020) Physiological and transcriptomic analyses of mulberry (Morus atropurpurea) response to cadmium stress. Ecotoxicol Environ Saf 205:111298
Danesh D, Aarons S, McGill GE, Young ND (1994) Genetic dissection of oligogenic resistance to bacterial wilt in tomato. Mol Plant Microbe Interact 7(4):464–471
Das S, Sultana KW, Chandra I (2021) Isolation and characterization of a plant growth-promoting bacterium Acinetobacter sp. SuKIC24 from in vitro-grown Basilicum polystachyon (L.) Moench. Curr Microbiol 78(8):2961–2969
DeBary A, Woronin M, Rothrock JT (1882) Morphologie und physiologie der pilze. Int J Plant Sci 7(7):81
Dong Z, Guo Y, Yu C, Zhixian Z, Rongli M, Deng W, Li Y, Hu X (2021) The dynamics in rhizosphere microbial communities under bacterial wilt resistance by mulberry genotypes. Arch Microbiol 203(3):1107–1121
Ercole TG, Savi DC, Adamoski D, Kava VM, Hungria M, Galli-Terasawa LV (2021) Diversity of maize (Zea mays L.) rhizobacteria with potential to promote plant growth. Braz J Microbiol 52(4):1807–1823
Fadiji AE, Babalola OO (2020) Metagenomics methods for the study of plant-associated microbial communities: a review. J Microbiol Methods 170:105860
Fu HZ, Marian M, Enomoto T, Hieno A, Ina H, Suga H, Shimizu M (2020) Biocontrol of tomato bacterial wilt by foliar spray application of a novel strain of endophytic Bacillus sp. Microbes Environ 35(4):ME20078
Girlich D, Bonnin RA, Proust A, Naas T, Dortet L (2021) Undetectable production of the VIM-1 Carbapenemase in an Atlantibacter hermannii clinical isolate. Front Microbiol 12:741972
Gond SK, Bergen MS, Torres MS, White JF Jr (2015) Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol Res 172:79–87
Hayward AC (1991) Biology and epidemiology of bacterial wilt caused by pseudomonas solanacearum. Annu Rev Phytopathol 29:65–87. https://doi.org/10.1146/annurev.py.29.090191.000433
Hiradate S, Yoshida S, Sugie H, Yada H, Fujii Y (2002) Mulberry anthracnose antagonists (iturins) produced by Bacillus amyloliquefaciens RC-2. Phytochemistry 61(6):693–698
Hong Lu, Zou WX, Meng JC, Jun Hu, Tan RX (2000) New bioactive metabolites produced by Colletotrichum sp., an endophytic fungus in Artemisia annua. Plant Sci 151:67–73
Huang F (1984) Introduction to the mulberry variety anti-cyanotropic 10. Guangdong Silk Newsl (03):12–17 (In China)
Kuźniar A, Włodarczyk K, Grządziel J, Goraj W, Gałązka A, Wolińska A (2020) Culture-independent analysis of an endophytic core microbiome in two species of wheat: Triticum aestivum L. (cv. ‘Hondia’) and the first report of microbiota in Triticum spelta L. (cv. ‘Rokosz’). Syst Appl Microbiol 43(1):126025
Lai W, Zeng X, Tan B, Wu G, Chen J, Guan W, Fan H (1979) Preliminary Identification of Pathogenic Bacteria of Mulberry Bacterial Wilt. Guangdong Silk Newsl (02):21–24 (In China)
Lai R, Ikram M, Li R, Xia Y, Yuan Q, Zhao W, Zhang Z, Siddique KHM, Guo P ( 2021) Identification of novel quantitative trait nucleotides and candidate genes for bacterial wilt resistance in Tobacco (Nicotiana tabacum L.) using genotyping-by-sequencing and multi-locus genome-wide association studies. Front Plant Sci 12:744175
Lee K, Yong D, Jeong SH, Chong Y (2011) Multidrug-resistant Acinetobacter spp.: increasingly problematic nosocomial pathogens. Yonsei Med J 52(6):879–891
Li B, Zhu F, Chen S, Lin Q, Qiu C, Zhu G (2010) Preliminary report on the survey of major diseases of mulberry in Guangxi. Guangxi Seric 47(02):23–26 (In China)
Lim SH, Choi CI (2019) Pharmacological properties of Morus nigra L. (Black Mulberry) as a promising nutraceutical resource. Nutrients 11(2):437
Litwińczuk W, Jacek B (2020) Micropropagation of Mountain Mulberry (Morus bombycis Koidz.) ‘Kenmochi’ on cytokinin-free medium. Plants (Basel) 9(11):1533
Liu Y, Vasiu S, Daughtrey ML, Filiatrault M (2020) First report of Dickeya dianthicola causing blackleg on New Guinea Impatiens (Impatiens hawkeri) in New York State, USA (published online ahead of print, 2020 Nov 17). Plant Dis. https://doi.org/10.1094/PDIS-09-20-2020-PDN
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plant 15:473–497
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27(5):824–834
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a Web browser. BMC Bioinform 12:385
Ou T, Xu WF, Wang F, Strobel G, Zhou ZY, Xiang ZH, Liu J, Xie J (2019) A microbiome study reveals seasonal variation in endophytic bacteria among different mulberry cultivars. Comput Struct Biotechnol J 17:1091–1100
Rana KL, Kour D, Kaur T, Devi R, Yadav AN, Yadav N, Dhaliwal HS, Saxena AK (2020) Endophytic microbes: biodiversity, plant growth-promoting mechanisms and potential applications for agricultural sustainability. Antonie Van Leeuwenhoek 113(8):1075–1107
Shin B, Park C, Park W (2020) Stress responses linked to antimicrobial resistance in Acinetobacter species. Appl Microbiol Biotechnol 104(4):1423–1435
Singh RK, Singh P, Guo DJ, Sharma A, Li DP, Li X, Verma KK, Malviya MK, Song XP, Lakshmanan P, Yang LT, Li YR (2021) Root-derived endophytic diazotrophic bacteria Pantoea cypripedii AF1 and Kosakonia arachidis EF1 promote nitrogen assimilation and growth in sugarcane. Front Microbiol 12:774707
Sun W, Xiong Z, Chu L, Li W, Soares MA, White JF Jr, Li H (2019) Bacterial communities of three plant species from Pb-Zn contaminated sites and plant-growth promotional benefits of endophytic Microbacterium sp. (strain BXGe71). J Hazard Mater 370:225–231
Tam DNH, Nam NH, Elhady MT, Tran L, Hassan OG, Sadik M, Tien PTM, Elshafei GA, Huy NT (2021) Effects of mulberry on the central nervous system: a literature review. Curr Neuropharmacol 19(2):193–219
Tian XL, Cao LX, Tan HM, Zeng QG, Jia YY, Han WQ, Zhou SN (2004) Study on the communities of endophytic fungi and endophytic actinomycetes from rice and their antipathogenic activities in vitro. World J Microbiol Biotechnol 20:303–309
Ulrich K, Kube M, Becker R, Schneck V, Ulrich A (2021) Genomic analysis of the endophytic Stenotrophomonas Strain 169 reveals features related to plant-growth promotion and stress tolerance. Front Microbiol 12:687463
Walterson AM, Stavrinides J (2015) Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev 39(6):968–984
Wang GF, Praphat K, Xie GL, Zhu B, Li B, Liu B, Zhou Q (2008) Bacterial wilt of mulberry (Morus alba) caused by Enterobacter cloacae in China. Plant Dis 92(3):483
Wood DE, Lu J, Langmead B (2019) Improved metagenomic analysis with Kraken 2. Genome Biol 20(1):257
Xiao L, Zhu Z, He K, Zhou M, Lin G (1980) Identification of resistance of mulberry varieties to mulberry bacterial wilt disease (continued). Guangdong Silk Newsl (03):8–10 (In China)
Xie J, Strobel GA, Feng T, Ren H, Mends MT, Zhou Z, Geary B (2015) An endophytic Coniochaeta velutina producing broad spectrum antimycotics. J Microbiol 53(6):390–397
Xie J, Shu P, Strobel G, Chen J, Wei J, Xiang Z, Zhou Z (2017) Pantoea agglomerans SWg2 colonizes mulberry tissues, promotes disease protection and seedling growth. Biol Control 113(1):9–17
Xu W, Wang F, Zhang M, Ou T, Wang R, Strobel G, Xiang Z, Zhou Z, Xie J (2019) Diversity of cultivable endophytic bacteria in mulberry and their potential for antimicrobial and plant growth-promoting activities. Microbiol Res 229:126328
Yang H (2018) Identification and molecular biology research of mulberry wilt pathogen. South China Agricultural University, Guangzhou
Yuan T, Luo L, Zhang X, Liu J (2022) Establishment of IMSA-LAMP detection method for Ralstonia solanacearum. J South China Agric Univ 43(04):89–98 ((In China))
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7(1–2):203–214
Zheng X, Zhu Y, Liu B, Yu Q, Lin N (2016) Rapid differentiation of Ralstonia solanacearum avirulent and virulent strains by cell fractioning of an isolate using high-performance liquid chromatography. Microb Pathog 90:84–92
Zhou Y, Yang H, Liu J (2021) Complete genome sequence of Enterobacter roggenkampii Strain KQ-01, isolated from bacterial wilt-resistant mulberry cultivar YS283. Plant Dis 105(3):688–690
Zhu B, Lou MM, Xie GL, Wang GF, Zhou Q, Wang F, Fang Y, Su T, Li B, Duan YP (2011) Enterobacter mori sp. nov., associated with bacterial wilt on Morus alba L. Int J Syst Evol Microbiol 61(Pt 11):2769–2774
Zhu F, Zhu G, Lin Q, Cen Z, Meng J, Chen X, Zeng Y, Qiu C, Ouyang Q (2014) Identification and evaluation of 76 mulberry hybrid combinations for resistance to cyanobacteria. Seric Sci 40(05):781–789 (In China)
Acknowledgements
This work was supported by the China Agriculture Research System (CARS-18-ZJ0304). We also acknowledge the assistance of other lab members who provided valuable support in this study, but are not listed as authors of this study.
Author information
Authors and Affiliations
Contributions
Ting YUAN designed the experiment, carried out the process of the experiment, analyzed the data and wrote the manuscript; Izhar Hyder QAZI assisted in data analysis and reviewed and edited the draft; Xueyin ZHANG, Jinhao LI, and Peijia YANG assisted in isolation of endophytes and data analysis; Jiping LIU provided project support, experimental design and edited the draft. All authors reviewed and approved the final version of manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, T., Qazi, I.H., Yang, P. et al. Analysis of endophytic bacterial flora of mulberry cultivars susceptible and resistant to bacterial wilt using metagenomic sequencing and culture-dependent approach. World J Microbiol Biotechnol 39, 163 (2023). https://doi.org/10.1007/s11274-023-03599-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03599-z