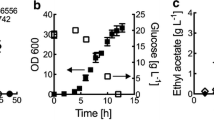
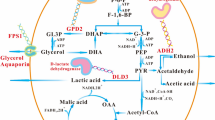
Abstract
Bioethanol, as a form of renewable and clean energy, has become increasingly important to the energy supply. One major obstacle in ethanol production is developing a high-capacity system. Existing approaches for regulating the ethanol production pathway are relatively insufficient, with nonspecific genetic manipulation. Here, we used CRISPR/Cas9 technology to disrupt the alcohol dehydrogenase (ADH) 2 gene via complete deletion of the gene and introduction of a frameshift mutation in the ADH2 locus. Sequencing demonstrated the accurate knockout of the target gene with 91.4% and near 100% targeting efficiency. We also utilized genome resequencing to validate the mutations in the ADH2 mutants targeted by various single-guide RNAs. This extensive analysis indicated the mutations in the CRISPR/Cas9-engineered strains were homozygous. We applied the engineered Saccharomyces cerevisiae strains for bioethanol production. Results showed that the ethanol yield improved by up to 74.7% compared with the yield obtained using the native strain. This work illustrates the applicability of this highly efficient and specific genome engineering approach to promote the improvement of bioethanol production in S. cerevisiae via metabolic engineering. Importantly, this study is the first report of the disruption of a target gene, ADH2, in S. cerevisiae using CRISPR/Cas9 technology to improve bioethanol yield.





Similar content being viewed by others
Abbreviations
- CRISPR:
-
Clustered regularly interspaced short palindromic repeat
- PAM:
-
Protospacer adjacent motif
- sgRNA:
-
Single-guide RNA
- DSB:
-
Double-strand break
- OD600:
-
Optical density at 600 nm
- PCR:
-
Polymerase chain reaction
- ADH:
-
Alcohol dehydrogenase
- GC:
-
Gas chromatography
- WT:
-
Wild type
- TALEN:
-
Transcription activator-like effector nuclease
- DSB:
-
Double strand break
- HR:
-
Homologous recombination
References
Balat M, Balat H (2009) Recent trends in global production and utilization of bio-ethanol fuel. Appl Energy 86:2273–2282
Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, Si T, Zhao H (2015) Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol 4:585–594
Baré G, Swiatkowski T, Moukil A, Gerday C, Thonart P (2002) Purification and characterization of a microbial dehydrogenase. Appl Biochem Biotechnol 98:415–428
Beier DR, Sledziewski A, Young ET (1985) Deletion analysis identifies a region, upstream of the ADH2 gene of Saccharomyces cerevisiae, which is required for ADR1-mediated derepression. Mol Cell Biol 5:1743–1749
Biot-Pelletier D, Martin VJ (2016) Seamless site-directed mutagenesis of the Saccharomyces cerevisiae genome using CRISPR-Cas9. J Biol Eng 10:6
Blandino A, Caro I, Cantero D (1997) Comparative study of alcohol dehydrogenase activity in flor yeast extracts. Biotechnol Lett 19:651–654
Boettcher M, McManus MT (2015) Choosing the right tool for the job: RNAi, TALEN, or CRISPR. Mol Cell 58:575–585
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chin YW, Kang WK, Jang HW, Turner TL, Kim HJ (2016) CAR1 deletion by CRISPR/Cas9 reduces formation of ethyl carbamate from ethanol fermentation by Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 43:1517–1525
Ciriacy M (1975) Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol Gen Genet 138:157–164
Ciriacy M (1979) Isolation and characterization of further cis- and trans-acting regulatory elements involved in the synthesis of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet 176:427–431
de Smidt O, du Preez JC, Albertyn J (2008) The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Res 8:967–978
de Smidt O, du Preez JC, Albertyn J (2012) Molecular and physiological aspects of alcohol dehydrogenases in the ethanol metabolism of Saccharomyces cerevisiae. FEMS Yeast Res 12:33–47
Demeke MM, Dietz H, Li Y, Foulquié-Moreno MR, Mutturi S, Deprez S, Abt D, Bonini T, Liden BM, Dumortier G, Verplaetse F, Boles A, Thevelein E, J.M (2013) Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6:89
DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343
Finnigan GC, Thorner J (2016) mCAL: a new approach for versatile multiplex action of Cas9 using one sgRNA and loci flanked by a programmed target sequence. G3 6:2147–2156
Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:38–41
He W, Ye S, Xue T, Xu S, Li W, Lu J, Cao L, Ye B, Chen Y (2014) Silencing the glycerol-3-phosphate dehydrogenase gene in Saccharomyces cerevisiae results in more ethanol being produced and less glycerol. Biotechnol Lett 36:523–529
Horwitz AA, Walter JM, Schubert MG, Kung SH, Hawkins K, Platt DM, Hernday AD, Mahatdejkul-Meadows T, Szeto W, Chandran SS, Newman JD (2015) Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst 1:88–96
Jakociunas T, Bonde I, Herrgard M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD (2015a) Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 28:213–222
Jakociunas T, Rajkumar AS, Zhang J, Arsovska D, Rodriguez A, Jendresen CB, Skjodt ML, Nielsen AT, Borodina I, Jensen MK, Keasling JD (2015b) CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. ACS Synth Biol 4:1226–1234
Jessop-Fabre MM, Jakociunas T, Stovicek V, Dai Z, Jensen MK, Keasling JD, Borodina I (2016) EasyClone-MarkerFree: a vector toolkit for marker-less integration of genes into Saccharomyces cerevisiae via CRISPR-Cas9. Biotechnol J 11:1110–1117
Kang HS, Charlop-Powers Z, Brady SF (2016) Multiplexed CRISPR/Cas9- and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Synth Biol 5:1002–1010
Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, Shumaker T, Wyrick JJ (2015) New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32:711–720
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Looke M, Kristjuhan K, Kristjuhan A (2011) Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques 50:325–328
Lutstorf U, Megnet R (1968) Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. I. Physiological control of ADH-2 and properties of ADH-2 and ADH-4. Arch Biochem Biophys 126:933–944
Manochio C, Andrade BR, Rodriguez RP, Moraes BS (2017) Ethanol from biomass: a comparative overview. Renew Sustain Energy Rev 80:743–755
Naito Y, Hino K, Bono H, Ui-Tei K (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics (Oxford England) 31:1120–1123
Norhayati Y, Shukuri MAM, Bakar SA, Aini ARN (2016) Effects of glucose, ethanol and acetic acid on regulation of ADH2 gene from Lachancea fermentati. Peerj 4(12):e1751
Rastogi M, Shrivastava S (2017) Recent advances in second generation bioethanol production: an insight to pretreatment, saccharification and fermentation processes. Renew Sustain Energy Rev 80:330–340
Reis VC, Nicola AM, de Souza Oliveira Neto O, Batista VD, de Moraes LM, Torres FA (2012) Genetic characterization and construction of an auxotrophic strain of Saccharomyces cerevisiae JP1, a Brazilian industrial yeast strain for bioethanol production. J Ind Microbiol Biotechnol 39:1673–1683
Ronda C, Maury J, Jakociunas T, Jacobsen SA, Germann SM, Harrison SJ, Borodina I, Keasling JD, Jensen MK, Nielsen AT (2015) CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microbiol Biotechnol 14:97
Russell DW, Smith M, Williamson VM, Young ET (1983) Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem 258:2674–2682
Sasano Y, Nagasawa K, Kaboli S, Sugiyama M, Harashima S (2016) CRISPR-PCS: a powerful new approach to inducing multiple chromosome splitting in Saccharomyces cerevisiae. Sci Rep 6:30278
Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH (2009) JBrowse: a next-generation genome browser. Genome Res 19:1630–1638
Soni KA, Lu L, Jesudhasan PR, Hume ME, Pillai SD (2008) Influence of autoinducer-2 (AI-2) and beef sample extracts on E. coli O157:H7 survival and gene expression of virulence genes yadK and hhA. J Food Sci 73:M135–M139
Steyn L, Du Preez J, de Smidt O, Albertyn J (2015) The use of multiple deletion mutants for determining the physiological role of alcohol dehydrogenase isozymes in Saccharomyces cerevisiae. https://doi.org/10.13140/RG.2.1.5014.8324
Tsarmpopoulos I, Gourgues G, Blanchard A, Vashee S, Jores J, Lartigue C, Sirand-Pugnet P (2016) In-yeast engineering of a bacterial genome using CRISPR/Cas9. ACS Synth Biol 5:104–109
Wills C (1976) Production of yeast alcohol dehydrogenase isoenzymes by selection. Nature 261:26–29
Xue T, Chen D-C, Liu Y, Su X-M, Lin R-H, Chen Y-Q (2015) Anthocyanin antisense interference of ethanol dehydrogenase II in Saccharomyces cerevisiae and its influence on ethanol fermentation. Mod Food Sci Technol. https://doi.org/10.13982/j.mfst.1673-9078.2015.11.022
Ye W, Zhang W, Liu T, Tan G, Li H, Huang Z (2016) Improvement of ethanol production in Saccharomyces cerevisiae by high-efficient disruption of the ADH2 gene using a novel recombinant TALEN vector. Front Microbiol 7:1067
Zabed H, Sahu JN, Suely A, Boyce AN, Faruq G (2017) Bioethanol production from renewable sources: current perspectives and technological progress. Renew Sustain Energy Rev 71:475–501
Acknowledgements
We thank Liangyi Zhou for help on verification of ADH2 mutants, Heqi Shao for advice on production of enzymatic extract. We thank Nature Research Editing Service (Order ID: 343TY1W5) for editing the English text of a draft of this manuscript.
Funding
This work was supported by the Natural Science Foundation of Fujian Province (Grant No. 2017J01622), the National Sugar Crop Reserach System (Grant No. CARS-170501), and the Education Department Project of Fujian Province (Grant No. JAT160114).
Author information
Authors and Affiliations
Contributions
Ting Xue, Kui Liu and Youqiang Chen designed the project; Kui Liu carried out the experiments; Kui Liu, Duo Chen, Xue Yuan and Jingping Fang analyzed the GC and genome resequencing data; Kui Liu wrote the manuscript; Ting Xue, Jingping Fang, Luqiang Huang, Youqiang Chen, Wenjin He edited the manuscript; All authors read and approved the final manuscript.
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xue, T., Liu, K., Chen, D. et al. Improved bioethanol production using CRISPR/Cas9 to disrupt the ADH2 gene in Saccharomyces cerevisiae. World J Microbiol Biotechnol 34, 154 (2018). https://doi.org/10.1007/s11274-018-2518-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-018-2518-4