Abstract
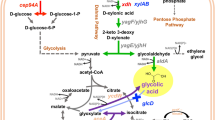
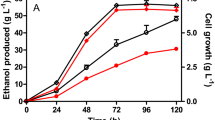
Anaerobic homofermentative production of reduced products requires additional reducing power (NADH and/or NADPH) output from glucose catabolism. Previously, with an anaerobically expressed pyruvate dehydrogenase operon (aceEF-lpd), we doubled the reducing power output to four NADH per glucose (or 1.2 xylose) catabolized anaerobically, which satisfied the NADH requirement to establish a non-transgenic homoethanol pathway (1 glucose or 1.2 xylose ⇒ 2 acetyl-CoA + 4 NADH ⇒ 2 ethanol) in the engineered strain, Escherichia coli SZ420 (∆frdBC ∆ldhA ∆ackA ∆focA-pflB ∆pdhR::pflBp6-pflBrbs-aceEF-lpd). In this study, E. coli SZ420 was further engineered for reduction of xylose to xylitol by (1) deleting the alcohol dehydrogenase gene (adhE) to divert NADH from the ethanol pathway; (2) deleting the glucose-specific PTS permease gene (ptsG) to eliminate catabolite repression and allow simultaneous uptake of glucose and xylose; (3) cloning the aldose reductase gene (xylI) of Candida boidinii to reduce xylose to xylitol. The resulting strain, E. coli AI05 (pAGI02), could in theory simultaneously uptake glucose and xylose, and utilize glucose as a source of reducing power for the reduction of xylose to xylitol, with an expected yield of four xylitol for each glucose consumed (YRPG = 4) under anaerobic conditions. In resting cell fermentation tests using glucose and xylose mixtures, E. coli AI05 (pAGI02) achieved an actual YRPG value of ~3.6, with xylitol as the major fermentation product and acetate as the by-product.


Similar content being viewed by others
References
Akinterinwa O, Cirino PC (2011) Anaerobic obligatory xylitol production in Escherichia coli strains devoid of native fermentation pathways. Appl Environ Microbiol 77(2):706–709
Alam KY, Clark DP (1989) Anaerobic fermentation balance of Escherichia coli as observed by in vivo nuclear magnetic resonance spectroscopy. J Bacteriol 171(11):6213–6217
Aristidou A, Penttila M (2000) Metabolic engineering applications to renewable resource utilization. Curr Opin Biotech 11(2):187–198
Berríos-Rivera S (2002) Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD+-dependent formate dehydrogenase. Metab Eng 4(3):217–229
Berríos-Rivera S, Bennett GN, San K (2002) The effect of increasing NADH availability on the redistribution of metabolic fluxes in Escherichia coli chemostat cultures. Metab Eng 4(3):230–237
Causey TB, Zhou S, Shanmugam KT, Ingram LO (2003) Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: homo-acetate production. Proc Natl Acad Sci USA 100:825–832
Chang DE, Shin S, Rhee JS, Pan JG (1999) Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J Bacteriol 181:6656–6663
Chin JW, Khankal R, Monroe CA, Maranas CD, Cirino PC (2009) Analysis of NADPH supply during xylitol production by engineered Escherichia coli. Biotech Bioeng 102(1):209–220
Cirino PC, Chin JW, Ingram LO (2006) Engineering Escherichia coli for xylitol production from glucose-xylose mixtures. Biotechnol Bioeng 95(6):1167–1176
Clark DP (1989) The fermentation pathways of Escherichia coli. FEMS Microbiol Rev 63:223–234
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
De Graef MR, Alexeeva S, Snoep JL, Mattos MJTD (1999) The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J Bacteriol 181:2351–2357
Gabor E, Gohler AK, Kosfeld A, Staab A, Kremling A, Jahreis K (2011) The phosphoenolpyruvate-dependent glucose-phosphotransferase system from Escherichia coli K-12 as the center of a network regulating carbohydrate flux in the cell. Eur J Cell Biol 90(9):711–720
Granstrom TB, Izumori K, Leisola M (2007) A rare sugar xylitol. Part I: the biochemistry and biosynthesis of xylitol. Appl Microbiol Biotechnol 74(2):277–291
Hernandez-Montalvo V, Valle F, Bolivar F, Gosset G (2001) Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl Microbiol Biotech 76(1–2):186–191
Jeffries TW (1983) Utilization of xylose by bacteria, yeasts, and fungi. Adv Biochem Eng Biotechnol 27:1–32
Kang MH, Ni H, Jeffries TW (2003) Molecular characterization of a gene for aldose reductase (CbXYL1) from Candida boidinii and its expression in Saccharomyces cerevisiae. Appl Biochem Biotechnol 105–106:265–276
Leonardo MR, Dailly Y, Clark DP (1996) Role of NAD regulating the adhE gene of Escherichia coli. J Bacteriol 178:6013–6018
Miller JH (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Press, Cold Spring Harbor
Nichols NN, Dien BS, Bothast RJ (2001) Use of catabolite repression mutants for fermentation of sugar mixtures to ethanol. Appl Microbiol Biotech 56(1–2):120–125
Posfai G, Koob MD, Kirkpatrick HA, Blattner FC (1997) Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157: H7 genome. J Bacteriol 179:4426–4428
Riondet C, Cachon R, Wache T, Alcaraz G, Divies C (2000) Extracellular oxidoreduction potential modifies carbon and electron flow in Escherichia coli. J Bacteriol 182(620):626
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Press, Cold Spring Harbor
Suzuki T, Yokoyama SI, Kinoshita Y, Yamada H, Hatsu M, Takamizawa K, Kawai K (1999) Expression of xylA gene encoding for d-xylose reductase of Candida tropicalis and production of xylitol in Escherichia coli. J Biosci Bioeng 87(3):280–284
Vandeska E, Amartey S, Kuzmanova Jeffries TW (1995) Effects of environmental conditions on production of xylitol by Candida boidinii. World J Microbiol Biotechnol 11(2):213–218
Woodyer R, Simurdiak M, van der Donk WA, Zhao HM (2005) Heterologous expression, purification, and characterization of a highly active xylose reductase from Neurospora crassa. Appl Environ Microbiol 71:1642–1647
Zhou S, Causey TB, Hasona A, Shanmugam KT, Ingram LO (2003) Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol 69:399–407
Zhou S, Iverson AG, Grayburn WS (2008) Engineering a native homoethanol pathway in Escherichia coli B for ethanol production. Biotechnol Lett 30:335–342
Zhou S, Iverson AG, Grayburn WS (2010) Doubling the catabolic reducing power (NADH) output of Escherichia coli fermentation for the production of reduced products. Biotechnol Prog 26:45–51
Acknowledgments
The chromosomal DNA of Candida boidinii (NRRL Y17213) was a kind gift from Dr. Thomas Jefferies of the University of Wisconsin. This research was supported by Northern Illinois University, and the Hubei University of Technology and the Chutian Scholar Program, P.R. China.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Iverson, A., Garza, E., Zhao, J. et al. Increasing reducing power output (NADH) of glucose catabolism for reduction of xylose to xylitol by genetically engineered Escherichia coli AI05. World J Microbiol Biotechnol 29, 1225–1232 (2013). https://doi.org/10.1007/s11274-013-1285-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-013-1285-5