Abstract
Pandemic H1N1 2009 (pH1N1), influenza virus containing triple reassortant internal genes (TRIG) from avian, human, and swine influenza viruses emerged in 2009 as a highly infectious virus that was able to be transmitted from humans to pigs. During June 2010–May 2012, influenza virus surveillance was conducted in Thai pig population. Twenty-three samples (1.75 %) were successfully isolated from total of 1,335 samples. Interestingly, pH1N1 (7 isolates, 30.34 %), reassortant pH1N1 (rH1N1) (1 isolate, 4.35 %), Thai endemic H1N1 (enH1N1) (3 isolates, 13.04 %), reassortant H3N2 with pH1N1 internal genes (rH3N2) (9 isolates, 39.13 %), and reassortant H1N2 with pH1N1 internal genes (rH1N2) (3 isolates, 13.04 %) were found. It should be noted that rH1N1, rH1N2, and rH3N2 viruses contained the internal genes of pH1N1 virus having a TRIG cassette descendant from the North American swine lineage. Although all isolates in this study were obtained from mild clinically sick pigs, the viruses were still highly infective and possibly may play an important role in human–animal interfacing transmission. In addition, the TRIG cassette may have an influence on antigenic shift resulting in emergence of novel viruses, as seen in this study. Continuing surveillance of influenza A natural hosts, particularly in pigs is necessary.
Similar content being viewed by others
Introduction
Since April 2009, pandemic H1N1 2009 (pH1N1) influenza virus emerged and spread fast worldwide among the human population, including Thailand. The molecular analysis indicated that pH1N1 is a reassortant virus between the North American triple reassortant swine influenza virus (SIV) and the European avian-like SIV. It was the first report of the reassorted SIV between North American and Eurasian lineages containing neuraminidase (NA) and matrix (M) genes from European avian-like SIV and six other genes from North American triple reassortant virus [1]. The North American triple reassortant virus composes of polymerase basic 2 (PB2), polymerase acidic (PA) genes from avian origin, polymerase basic 1 (PB1) genes from human origin and hemagglutinin (HA), nucleoprotein (NP), and nonstructural (NS) genes from classic swine lineage. The triple reassortant internal gene (TRIG) cassette has a high tendency to pick up different surface proteins, and is able to continuingly survive in pigs and in humans. As a result, the genetic characteristic of North American SIVs has high variations after the introduction of the TRIG virus into the swine populations [2, 3].
Influenza virus circulates among different host species including wild birds, poultry, pigs, horses, and humans [4]. The viruses usually favorably binds to 2 specific receptors, sialic acid (SA) α2,3 found in the epithelial cells of the gastrointestinal tract of wild aquatic birds, and SA α2,6 found in the epithelial cells of human respiratory tract depending on the virus origin. Recent studies demonstrate that pigs have abundant expression of SA α2,6 receptors in the upper respiratory tract, but α2,3-linked SA receptors were only expressed in the lower respiratory tract with moderate intensity, similar to that in humans [5–7]. Hence, the Thai swine ecosystem may enhance the reassortant between human and pig viruses. In addition, cross-species transmission was occasionally found [8, 9] due to commingling among domestic species, and it is commonly practiced in the backyard farming systems, and in small to medium swine farms in most Asian countries, including Thailand.
Several months after pH1N1 emergence in humans, the virus was isolated from pigs not showing clinical signs, thus confirming human to pig transmission in many countries, including Canada, Norway, Italy, Hong Kong, and Thailand [10–14]. Subsequently, pH1N1 reassortant (rH1N1) viruses were reported in Hong Kong, Italy, German, Thailand, and USA [13, 15–17]. It should be noted that the Thai rH1N1 virus contained the NA gene from a Thai endemic SIV and other seven genes from pH1N1.
Retrospectively, genetic characterization of the Thai SIVs during 2000–2008 showed 3 major SIV subtypes (H1N1, H1N2, and H3N2) [18]. The endemic Thai H1N1 SIV had internal genes, and NA gene is closely related to the avian-like swine lineage of Eurasia, but HA gene is similar to classic swine H1N1. The H3 HA viruses were also divided into two clades. Clade A was similar to human H3N2 virus and clade B was similar to human-like swine SIV from European countries and Hong Kong. The N2 NA gene of Thai H3N2 subtype was divided into three groups; American/Asian SIV, European SIV, and Thai SIV. In contrast, H1N2 virus possesses the H1 HA gene from classic swine H1N1, but the N2 NA gene from European lineage [8, 18, 19]. It should be noted that the TRIG cassette swine influenza viruses had never been found in Thailand before 2009. It was not until the introduction of pH1N1 into the Thai pig population in early 2009 [12]. As a result, genetic surveillance of Thai SIV is important and necessary for disease control and prevention of the novel influenza viruses.
Materials and methods
Sample collection
A total of 1,314 nasal swabs and 21 lung tissues were collected from April 2010 to May 2012. The Thai influenza surveillance was performed in 28 pig farms in 13 provinces, in Northern, Northeastern, Eastern, and Central regions of the country. Nasal swabs were collected from 4–8, 12, 16, 20, 24-week-old pigs, gilts, and sows with noticeable respiratory signs, such as sneezing, coughing, conjunctivitis, and fever. Lung tissues were collected from 5 to 6-week-old necropsied pigs with lesions characterized by multifocal, dark, plum-colored consolidation at cranioventral areas, similar to Mycoplasma hyopneumoniae-infected lung. Other respiratory pathogens included porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2) were also tested in all samples.
Virus identification and isolation
Total RNA was extracted from nasal swabs or supernatant of lung tissue homogenate by using a commercial kit (NucleoSpin Extract Viral RNA Kit, Macherey-Nagel, Germany) as described by the manufacturer, and submitted for a routine reverse transcriptase polymerase chain reaction (RT-PCR) [20]. Primers specific to Matrix (M) gene (forward primer: 5′-TGATCTTCTTGAAAATTTGCAG-3′ and reverse primer: 5′-CGATGGTCATTTTGTCAACA-3′) were used and RT-PCR system was performed by using AccessQuick RT-PCR system (Promega, USA). Briefly, the cycling conditions started at 48 °C for 45 min, 94 °C for 3 min, and were followed by 35 cycles of denaturation (94 °C for 20 s), annealing (55 °C for 20 s), and extension (72 °C for 30 s). RT-PCR positive samples were subsequently inoculated into 9-day-old embryonated chicken eggs and MDCK cell line as described previously [21]. Isolated viruses were harvested within 2 days after inoculation for virus amplifying before genetic sequencing.
Virus gene sequencing and sequencing analysis
Viral RNA was extracted from harvested amniotic fluid or cell culture supernatant and cDNAs were synthesized by using ImProm-II Reverse Transcriptase (Promega, USA) and universal primer (5′-AGCAAAAGCAGG-3′), as described by the manufacturer. Then, PCRs were performed using specific primers designed for full length of eight genes of influenza A virus [22]. The PCR products were analyzed by using 1.2 % agarose gel electrophoresis and purified by using a commercial kit (Nucleospin Gel and PCR clean-up, Macherey–Nagel, Germany). DNA sequencing was carried out by 1st BASE company (Singapore) with specific primer sets. Blast analysis was carried out on NCBI. Sequence analysis was done by using MEGA 5.1 program [23]. The nucleotide sequences of Thai SIV isolates, human pH1N1, Thai endemic swine H1N1 (previously isolated in 2006–2009), North American swine, European swine and avian H1N1 viruses were included in H1 HAs and N1 NAs phylogenetic tree. In H3 HAs and N2 NAs phylogenetic trees, human H3N2, Thai endemic swine H3N2 (previously isolated in 2004–2005), North American swine, European swine and avian H3N2 viruses were included. All reference isolates were obtained from Genbank. The phylogenetic tree was constructed applying neighbor-joining algorithm by using MEGA 5.1 program.
Results
Virus identification and sequence analysis
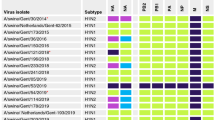
Out of the 1,335 samples (1,314 swabs and 21 lung tissues), only 23 samples (1.75 %) were positive for influenza A virus based on Matrix gene RT-PCR results. Either PRRSV or PCV2 or both were also detected in the collected samples (data not shown). All 23 positive samples from 4 to 8-week-old pigs were then isolated and amplified before sequencing. It should be noted that most of the positive samples were obtained during the seasonal flu epidemic from October to March in Thailand. These positive samples were collected from 11 farms (39.28 %, 11/28) located in the central area (Saraburi, Suphanburi, and Ratchaburi), the Eastern area (Chonburi and Chachoengsao), the Northeastern area (Nakorn Ratchasima, and Burirum), and the Northern area (Chaing Mai and Lumpoon) of Thailand. Detailed description and Genbank accession number of each isolate are shown in Table 1.
From a total of 23 RT-PCR positive samples, seven isolates (A/swine/Thailand/CU-RP1/2010, A/swine/Thailand/CU-RP3/2010, A/swine/Thailand/CU-SPN47/2010, A/swine/Thailand/CU-SPN65/2010, A/swine/Thailand/CU-PL63/2010, A/swine/Thailand/CU-PL65/2010, and A/swine/Thailand/CU-DP83/2010) were similar to pH1N1 virus. The sequence analysis revealed these swine pH1N1 viruses had all genes, including HA and NA genes closely related to the human pH1N1 virus (Table 2).
One rH1N1 (A/swine/Thailand/CU-SA433/2010) was re-isolated from the same farm, as previously reported [15]. This isolate contained PB2, PB1, PA, HA, NP, M, and NS genes of the pH1N1 virus, but the NA gene closely related to that of the endemic swine H1N1 viruses circulating in Thailand, Hong Kong, and European countries (Table 2).
Three isolates were closely related to the endemic Thai H1N1 (enH1N1) virus, previously isolated in 2004–2006 [18]. These three isolates included A/swine/Thailand/CU-SPL2/2010, A/swine/Thailand/CU-SPL4/2010, and A/swine/Thailand/CU-PS73/2010. The PB2, PB1, HA, NP, and NA genes showed 94–96 % identity to the Thai enH1N1 in 2000 (A/swine/Ratchaburi/NIAH1481/2000(H1N1) (Table 2), whereas, PA, M, and NS were 95–98 % identity to the Thai enH1N1 in 2004 (A/swine/Chonburi/NIAH977/2004(H1N1).
Interestingly, from 2011 onward, nine reassortant H3N2 viruses (rH3N2) and three reassortant H1N2 (rH1N2) viruses containing pH1N1 internal genes were found. The rH3N2 viruses included A/swine/Thailand/CU-CG43/2011, A/swine/Thailand/CU-CG45/2011, A/swine/Thailand/CU-CG48/2011, A/swine/Thailand/CU-CG51/2011, A/swine/Thailand/CU-CG55/2011, A/swine/Thailand/CU-P43/2012, A/swine/Thailand/CU-P53/2012, A/swine/Thailand/CU-BN53/2012, and A/swine/Thailand/CU-BN54/2012. These rH3N2 viruses had PB2, PB1, PA, NP, M, and NS genes similar to those of pH1N1 virus, but HA and NA genes of these viruses had 95 % identity to the previous Thai swine H3N2 isolate [A/swine/Ratchaburi/NIAH59/2004(H3N2)] (Table 2). The three rH1N2 viruses including (A/swine/Thailand/CU-CT43/2011, A/swine/Thailand/CU-CT63/2011, and A/swine/Thailand/CU-CT83/2011) were obtained from the same farm. PB2, PB1, PA, HA, NP, M, and NS genes were closely related to those of pH1N1 virus, but NA gene had 93 % identity to human H3N2 isolate [A/Stockholm/12/1988(H3N2)] and had only 88 % identity to the previous Thai swine isolate [A/swine/Ratchaburi/NIAH9426/2005(H3N2)] (Table 2).
Phylogenetic analysis
Phylogenetic tree of H1 HAs showed that HA gene of pH1N1, rH1N1, and rH1N2 viruses were grouped together with human pH1N1 isolates (A/California/04/2009 and A/Nonthaburi/102/2009), whereas, HA gene of enH1N1 viruses belonged to the North American classic swine lineage, similar to the 2000–2009 endemic H1N1 SIV in Thailand (A/swine/Thailand/CU-CB1/2006, A/swine/Thailand/CU-ST3/2009) (Fig. 1a).
The phylogenetic tree of H3 HAs is shown in Fig. 1b. All isolates in this study were clustered with endemic H3N2 viruses in Thailand during 2004–2005. These viruses were distant from avian, human and European swine lineages. In addition, the Thai rH3N2 isolates were distinguished from the 2011 reassortant H3N2 virus isolated from humans in North America (A/Indiana/08/2011 and A/West Virginia/06/2011).
The phylogenetic tree of N1 NAs is shown in Fig. 1c. NA gene of swine pH1N1 isolates was grouped with human pH1N1 virus. The N1 NAs of enH1N1 and rH1N1 viruses were grouped with endemic SIV from 2005 to 2009. Therefore, N1 NAs of pH1N1, enH1N1, and rH1N1 viruses were grouped together in European avian-like swine lineage and were distant from avian, human, and North American classic swine lineages.
The phylogenetic tree of N2 NAs showed that NA gene of Thai rH3N2 viruses clustered in the same branch with endemic Thai swine H3N2 isolated during 2004–2005. However, the rH3N2 viruses were distinguished from human, North American swine lineage and the 2011 reassortant human viruses. Interestingly, NA gene of the rH1N2 isolates was grouped in the same branch with a human virus isolated in 1988 (A/Stockholm/12/1988), which is different from the other N2 isolates in this study (Fig. 1d).
Patterns of all pH1N1 reassortant viruses (rH1N1, rH1N2 and rH3N2) were demonstrated in Fig. 2.
The schematic diagram of reassortant patterns of recent Thai pH1N1 reassortant viruses (rH1N1, rH1N2 and rH3N2). The rH1N1 picked the NA gene up from the endemic H1N1 and other 7 genes were similar to the pH1N1 virus. The rH3N2 contained 6 internal genes from pH1N1 and both envelope genes were from Thai endemic H3N2 previously circulated in Thai pig population in 2005. The rH1N2 picked the NA gene up from a human H3N2 virus and other 7 genes were similar to the pH1N1 virus
Molecular analysis of HA protein receptor-binding and antigenic site
The receptor binding site of H1 HAs was determined by amino acid position 204 and 239. The aspartic acid (D) at both positions conferred binding to human influenza virus receptor (SA α2,6), whereas, glutamic acid (E) and glycine (G) conferred binding to avian receptor (SA α2,3) [7, 24]. All swine isolates of pH1N1, rH1N1, two isolates of enH1N1 (A/swine/Thailand/CU-SPL2/2010 and A/swine/Thailand/CU-PS73/2010), and two isolates of rH1N2 (A/swine/Thailand/CU-CT43/2011 and A/swine/Thailand/CU-CT83/2011) posed D in both positions (Table 3). One enH1N1 (A/swine/Thailand/CU-SPL4/2010) and one rH1N2 (A/swine/Thailand/CU-CT63/2011) had D at amino acid position 204, but had G at position 239 (data not shown).
The H3 HAs receptor binding site was determined by amino acid position 242 and 244. The glutamine (Q) and G at position 242 and 244 conferred binding to Sialic acid (SA) α2,3 receptor, whereas, leucine (L) and serline (S) were compatible with SA α2,6 receptor. All H3 isolates in this study had L at position 242 and S at position 244 (Table 4).
The H1 HAs posed 4 antigenic sites; Sa, Sb, Ca, and Cb. The antigenic sites of swine pH1N1, rH1N1, and rH1N2 viruses were similar to the human pH1N1 virus (A/California/04/2009), but different from enH1N1 (A/swine/Thailand/CU-CB1/2006), European swine (A/swine/Belgium/1/1998) and human H1N1 lineage (A/Puerto Rico/8/1934) (Table 3). The cleavage site at amino acid position 339–350 was a typical of H1 subtype, and was found in all H1 isolates (Table 3). All H1 HAs isolates in this study had eight potential glycosylation sites similar to North American classic swine lineage. On the other hand, the European avian-like lineage had seven potential glycosylation sites (position 293–295 was not a glycosylation site) (data not shown).
The H3 HAs had five antigenic sites; A–E. The antigenic sites of rH3N2 isolates were similar to the Thai swine virus (A/swine/Ratchaburi/NIAH59/2004) and human H3N2 virus (A/Brisbane/8/1996) (Table 4). Human H3 HAs viruses had ten potential glycosylation sites. However, Thai rH3N2 isolates had eight potential glycosylation sites (amino acid position 149–151 and 160–162 were not glycosylation sites) (data not shown).
Molecular analysis of drug resistance on NA and M proteins
Oseltamivir-resistance is correlated with NA gene amino acid mutation E120V, H275Y, R293K, and N295S of N1 NAs and E119V, H274Y, R292K, and N294S of N2 NAs [5]. All N1 NAs and N2 NAs isolates in this study contained amino acid substitution at these four positions are considered as Oseltamivir-resistance.
Amantadine-resistance is involved with amino acid mutation L26F, V27A, A30T, S31N, and G34E of M gene [5]. All isolates in this study contained amino acid substitution S31N, but no amino acid mutation at 26F, 27A, 30T, and 34E.
Molecular analysis of virulence factors on PB2, PB1, and NS proteins
The virulence of influenza A virus may be associated with mutation of amino acid positions E627K and N701D of PB2, S66N of PB1-F2, and E92D of NS1 proteins [25]. PB2 protein of swine pH1N1, rH1N1, rH1N2, and rH3N2 isolates in this study contained E and D at amino acid position 627 and 701, respectively (indicating low or high virulence). In contrast, enH1N1 isolates had E and Aspargine (N) at 627 and 701 position of PB2 protein. All pH1N1 and the reassortant isolates had stop codon inside the PB1-F2 protein. The enH1N1 isolates had no amino acid substitution at amino acid position 66 of PB1-F2 protein. All isolates in this study showed no amino acid mutation at position 92 of NS1 protein.
Discussion
The surveillance data indicated that pH1N1 viruses were mainly found among Thai pigs during year 2010, after the introduction in 2009. Most viruses were recovered from medium to large swine farms with no animal imported from outsides, indicating that pH1N1 was successfully transmitted from humans to pigs, becoming dominant and later endemic in late 2010 and early 2011 in the Thai swine population. These findings correlated well with the human influenza virus surveillance results in Thailand [26] showing that pH1N1 was the most prevalent virus found in 2010, and later the H3N2 virus replaced and became the dominant virus in late 2011.
Interestingly, all isolates in this study were obtained from the nursery pigs indicating that those isolated viruses became endemic in the swine population, and nursery pigs are susceptible to SIV-infection due to the declining of maternal-derived antibody. It should be noted that the Thai endemic SIVs (endemic H1N1, H1N2, and H3N2) were occasionally found in mid 2010 (the first 6 months of the surveillance) and never been isolated again. Later, pH1N1 virus appeared to be the most dominant virus replacing Thai endemic SIVs. However, in 2011–2012, the rH3N2 viruses became the major virus population in Thai pigs, whereas, the rH1N2 viruses were found only in one swine farm at the end of 2011. Interestingly, the rH1N2 NA gene was grouped in the same branch with a human virus isolated in 1988 (A/Stockholm/12/1988) different from the other endemic H3N2 or rH3N2 in Thailand implying that the rH3N2 virus might obtain the N2 gene from a human virus. Again, swine workers and veterinarians, when sick with flu-like symptoms should not come into contact with pigs in order to reduce the chance of introducing new virus genes into the pig population.
It should be noted that the novel reassortant H3N2 virus was isolated from humans in the United State since June 2011 [9]. The virus PB2, PB1, PA, HA, NP, NA, and NS genes were closely related to the TRIG SIV H3N2 virus, but its M gene was closely related to pH1N1 virus. Thus, the novel H3N2 virus found in the US did not relate to the rH3N2 isolates in this study.
Since 2009, variation of reassortant viruses was frequently found in the Thai pig population after the pH1N1 emergence in the Thai pig population. The pH1N1 virus contains TRIG cassette and influences the virus genetic shift or reassortant, as previously demonstrated in North America [3, 27]. The TRIG cassette virus was introduced into the North American swine population in 1988 inducing variation of genetic reassortant of SIVs [27]. From 1998 to 2008, at least 7–8 types of influenza A viruses were isolated from the North American swine population. It should be noted that during 1930–1997 only one classic SIV was isolated and was genetically stable for almost 60 years [27]. Similarly, Thai SIV genetic characteristics had been stable before the introduction of the pH1N1 or TRIG cassette virus in 2009. The rH3N2 isolates found in this study demonstrated a similar situation already mentioned above, happening in Thailand. The rH3N2 viruses were isolated from two swine farms located 822 km apart, and those farms have not imported pigs from outsources. This information suggests that the TRIG cassette viruses frequently acquire new envelop proteins for escaping the host immune response [27].
In this study, all the Thai reassortant viruses containing the TRIG cassette were the results of rapid antigenic drift and shift. These HA and NA genes were derived from genetic compositions of endemic SIVs from Thai pigs (except for rH1N2 isolates) [8, 18, 19]. This present information pointed to the fact that pH1N1, enH1N1, and Thai endemic H3N2 viruses were co-circulating within the Thai pig population. Since pH1N1 virus was the most dominate viruses during late 2010–early 2011, numbers of pH1N1 reassortant viruses were later isolated in 2011–2012.
Most of the swine viruses isolated in this study possessed receptor-binding site compatible with human receptor (SA α2,6) [7–25, 28, 29]. In addition, pH1N1 virus contains TRIG cassette making the virus become highly infective among these species. Cross-species transmission between pigs and humans could not be avoided, particularly in the swine-exposed population. Veterinarians and swine workers have a high exposure risk to the SIV [30]. A good example of interspecies transmission was the novel reassortant influenza H3N2 virus in the United State in 2011. The virus was isolated from 12 people, but only six people had a history of coming in contact with swine, suggesting probably human-to-human transmission route [9]. This possibility could occur in Thailand, since people do not pay much attention to flu or flu-like symptoms when working closely to swine. This ignorance may facilitate the novel reassortant virus to spread into human communities or vice versa.
Amino acid substitution of NA of all Thai swine isolates in this study showed Oseltamivir-resistance. Amino acid substitution (S31N) of M genes was also able to cause Amantadine-resistance [31]. However, pH1N1 and its reassortant viruses were possibly of low virulence, according to no amino acid substitution at position 627, 701 of PB2, 92 of NS1 protein and non-translation of PB1-F2 protein [5, 25]. Additionally, the pathogenesis of pH1N1 and its reassortant virus found in Thai pigs was conducted, and found only mild clinical signs in experimentally infected nursery pigs [32]. The endemic H1N1 virus might cause more severe clinical signs because of having amino acid substitution N701D of PB2 protein, as shown in the previous pathogenesis study of a local Thai SIV [21].
In summary, pH1N1 virus and its TRIG cassette reassortant viruses have recently been established in the Thai swine population [33]. The TRIG cassette viruses rapidly drive antigenic drift and shift causing various reassortant viruses of the pH1N1 origin. Two major concerns include having increased numbers of novel genetic reassortant viruses and public health consequences when having highly pathogenic reassortant viruses. Definitely, genetic characteristic of future Thai SIV will be more variable. In addition, genetic variations of the future Thai SIVs may affect the swine herd health immunity differently from the endemic viruses, and may cause economic loss in the Thai swine industry more or less. Public health awareness should be a major focus due to evidence of interspecies transmission among human and pig populations, since pigs and humans do have similar expression of SA receptors in the respiratory tract [6, 7]. The TRIG cassette viruses were likely to acquire new HA and NA genes, resulting in the emergence of novel reassortment viruses and can probably successfully replicate in humans. In addition, during 2003–2004, avian influenza H5N1 virus emerged in Thailand, and caused 12 human deaths with limited human-to-human transmission [34, 35]. Although no reassortant evidence between the TRIG cassette virus and the avian H5N1 virus was previously found, this particular novel virus could be reassorted from the commingling environment among domestic species commonly seen in the backyard farming system. The combination of the high infectivity TRIG cassette and high virulent avian H5N1 viruses might possibly cause a dangerous pandemic threat among human population in the future. Thus, continuous influenza surveillance is necessary, particularly in swine.
References
R.J. Garten, C.T. Davis, C.A. Russell, B. Shu, S. Lindstrom, A. Balish, W.M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C.B. Smith, S.L. Emery, M.J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D.F. Burke, R.A. Fouchier, C. Pappas, C.M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C.A. Myers, D. Faix, P.J. Blair, C. Yu, K.M. Keene, P.D. Dotson Jr, D. Boxrud, A.R. Sambol, S.H. Abid, K. St George, T. Bannerman, A.L. Moore, D.J. Stringer, P. Blevins, G.J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H.F. Guevara, E.A. Belongia, P.A. Clark, S.T. Beatrice, R. Donis, J. Katz, L. Finelli, C.B. Bridges, M. Shaw, D.B. Jernigan, T.M. Uyeki, D.J. Smith, A.I. Klimov, N.J. Cox, Science 325, 197–201 (2009)
W. Ma, A.L. Vincent, K.M. Lager, B.H. Janke, S.C. Henry, R.R. Rowland, R.A. Hesse, J.A. Richt, Virus Genes 40, 28–36 (2010)
A.L. Vincent, S.L. Swenson, K.M. Lager, P.C. Gauger, C. Loiacono, Y. Zhang, Vet. Microbiol. 137, 51–59 (2009)
H.L. Forrest, R.G. Webster, Res. Rev. 11, 3–18 (2010)
C.F. Arias, M. Escalera-Zamudio, M. de Los, D. Soto-Del Río, A.G. Cobián-Güemes, P. Isa, S. López, Arch. Med. Res. 40, 643–654 (2009)
S.G. Van Poucke, J.M. Nicholls, H.J. Nauwynck, K. Van Reeth, Virol J. 7(38), 1–14 (2010)
M. Imai, Y. Kawaoka, Curr. Opin. Virol. 2, 160–167 (2012)
N. Komadina, V. Roque, P. Thawatsupha, J. Rimando-Magalong, S. Waicharoen, E. Bomasang, P. Sawanpanyalert, M. Rivera, P. Iannello, A.C. Hurt, I.G. Barr, Virus Genes 35, 161–165 (2007)
S. Lindstrom, R. Garten, A. Balish, B. Shu, S. Emery, L. Berman, N. Barnes, K. Sleeman, L. Gubareva, J. Villanueva, A. Klimov, Emerg. Infect. Dis. 18, 834–837 (2012)
M. Hofshagen, B. Gjerset, C. Er, A. Tarpai, E. Brun, B. Dannevig, T. Bruheim, I.G. Fostad, B. Iversen, O. Hungnes, B. Lium, Euro. Surveill. 14, 687–689 (2009)
A. Moreno, I. Barbieri, E. Sozzi, A. Luppi, D. Lelli, G. Lombardi, M.G. Zanoni, P. Cordioli, Vet. Microbiol. 138, 361–367 (2009)
D. Sreta, S. Tantawet, S.N. Na Ayudhya, A. Thontiravong, M. Wongphatcharachai, J. Lapkuntod, N. Bunpapong, R. Tuanudom, S. Suradhat, L. Vimolket, Y. Poovorawan, R. Thanawongnuwech, A. Amonsin, P. Kitikoon, Emerg. Infect. Dis. 16(10), 1587–1590 (2010)
D. Vijaykrishna, L.L. Poon, H.C. Zhu, S.K. Ma, O.T. Li, C.L. Cheung, G.J. Smith, J.S. Peiris, Y. Guan, Science 328, 1529 (2010)
H.M. Weingartl, Y. Berhane, T. Hisanaga, J. Neufeld, H. Kehler, C. Emburry-Hyatt, K. Hooper-McGreevy, S. Kasloff, B. Dalman, J. Bystrom, S. Alexandersen, Y. Li, J.J. Pasick, Virology 84, 2245–2256 (2010)
P. Kitikoon, D. Sreta, S. Nuntawan Na Ayudhya, M. Wongphatcharachai, J. Lapkuntod, D. Prakairungnamthip, N. Bunpapong, S. Suradhat, R. Thanawongnuwech, A. Amonsin, Virus Genes 43, 1–5 (2011)
A. Moreno, L. Di Trani, S. Faccini, G. Vaccari, D. Nigrelli, M.B. Boniotti, E. Falcone, A. Boni, C. Chiapponi, E. Sozzi, P. Cordioli, Vet. Microbiol. 149, 472–477 (2011)
E. Starick, E. Lange, S. Fereidouni, C. Bunzenthal, R. Hoveler, A. Kuczka, E. Grosse Beilage, H.P. Hamann, I. Klingelhofer, D. Steinhauer, T. Vahlenkamp, M. Beer, T. Harder, J. Gen. Virol. 92, 1184–1188 (2011)
S. Chutinimitkul, N. Thippamom, S. Damrongwatanapokin, S. Payungporn, R. Thanawongnuwech, A. Amonsin, P. Boonsuk, D. Sreta, N. Bunpong, R. Tantilertcharoen, P. Chamnanpood, S. Parchariyanon, A. Theamboonlers, Y. Poovorawan, Arch. Virol. 153, 1049–1056 (2008)
N. Takemae, S. Parchariyanon, S. Damrongwatanapokin, Y. Uchida, R. Ruttanapumma, C. Watanabe, S. Yamaguchi, T. Saito, Influenza Other Respir. Viruses 2, 181–189 (2008)
S. Payungporn, S. Chutinimitkul, A. Chaisingh, S. Damrongwantanapokin, C. Buranathai, A. Amonsin, A. Theamboonlers, Y. Poovorawan, J. Virol. Methods 131, 143–147 (2006)
D. Sreta, R. Kedkovid, S. Tuamsang, P. Kitikoon, R. Thanawongnuwech, Virol J. 6, 1–11 (2009)
E. Hoffman, J. Stech, Y. Guan, R.G. Webster, D.R. Perez, Arch. Virol. 146, 2275–2289 (2001)
K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei, S. Kumar, Mol. Biol. Evol. 28, 2731–2739 (2011)
M. Igarashi, K. Ito, R. Yoshida, D. Tomabechi, H. Kida, A. Takada, PLoS ONE 5, e8553 (2010)
G. Neumann, T. Noda, Y. Kawaoka, Nature 459, 931–939 (2009)
The global influenza surveillance and response system, Laboratory surveillance information. http://www.who.int/flunet. Accessed 26 Oct 2012
A.L. Vincent, W. Ma, K.M. Lager, B.H. Janke, J.A. Richt, Adv. Virus Res. 72, 127–154 (2008)
J.J. Skehel, D.C. Wiley, Annu. Rev. Biochem. 69, 531–569 (2000)
Y. Suzuki, Biol. Pharm. Bull. 28, 399–408 (2005)
P. Kitikoon, D. Sreta, R. Tuanudom, A. Amonsin, S. Suradhat, K. Oraveerakul, Y. Poovorawan, R. Thanawongnuwech, Vet. Microbiol. 148, 413–418 (2011)
K. Das, J.M. Aramini, L.C. Ma, R.M. Krug, E. Arnold, Nat. Struct. Biol. 17, 530–538 (2010)
N. Charoenvisal, J. Keawcharoen, D. Sreta, S. Tantawet, S. Jittimanee, J. Arunorat, A. Amonsin, R. Thanawongnuwech, Virol J. 10, 88 (2013)
Y. Hiromoto, S. Parchariyanon, N. Ketusing, P. Netrabukkana, T. Hayashi, T. Kobayashi, N. Takemae, T. Saito, Virus Res. 169, 175–181 (2012)
J. Keawcharoen, A. Amonsin, K. Oraveerakul, S. Wattanodorn, T. Papravasit, S. Karnda, K. Lekakul, R. Pattanarangsan, S. Noppornpanth, R.A.M. Fouchier, A.D.M.E. Osterhaus, S. Payungporn, A. Theamboonlers, Y. Poovorawan, Acta Virol. 49, 277–280 (2005)
World Health Organization, Cumulative number of confirmed human cases of avian influenza A (H5N1) reported to WHO. http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/index.html. Accessed 1 Mar 2013
Acknowledgments
This study has been funded in part with federal funds from the National Institute of Allergy and Infectious Disease, National Institute of Health, Department of Health and Human Services, under Contract No. HHSN266200700007c. Its contents are solely the responsibility of the authors, and do not necessarily represent the official views of the NIH. Additionally, this study was supported by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission (HR1160A and HR1155A), and continuous Chulalongkorn University research funding of CHULAR2 (Health cluster) and “Integrated Innovation Academic Center: IIAC” Chulalongkorn University Centenary Academic Development Project and the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund) for Dr. Nataya Charoenvisal PhD program. Finally, the authors would like to express their sincere thanks to Dr. Aunyaratana Thontiravong, Mr. Cherdpong Poophonpan, Miss Wikanda Tunterak, Miss Duangduean Prakairungnamthip, and Miss Ranida Tuanudom for their technical assistance, and Dr. Neel Aziz for editing the revised manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Charoenvisal, N., Keawcharoen, J., Sreta, D. et al. Genetic characterization of Thai swine influenza viruses after the introduction of pandemic H1N1 2009. Virus Genes 47, 75–85 (2013). https://doi.org/10.1007/s11262-013-0927-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-013-0927-x