Abstract
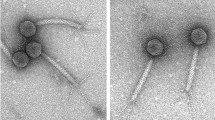
“Natto”, considered a traditional food, is made by fermenting boiled soybeans with Bacillus subtilis (natto), which is a natto-producing strain related to B. subtilis. The production of natto is disrupted by phage infections of B. subtilis (natto); hence, it is necessary to control phage infections. PM1, a phage of B. subtilis (natto), was isolated during interrupted natto production in a factory. In a previous study, PM1 was classified morphologically into the family Siphoviridae, and its genome, comprising approximately 50 kbp of linear double-stranded DNA, was assumed to be circularly permuted. In the present study, the complete nucleotide sequence of the PM1 genomic DNA of 50,861 bp (41.3 %G+C) was determined, and 86 open reading frames (ORFs) were deduced. Forty-one ORFs of PM1 shared similarities with proteins deduced from the genome of phages reported so far. Twenty-three ORFs of PM1 were associated with functions related to the phage multiplication process of gene control, DNA replication/modification, DNA packaging, morphogenesis, and cell lysis. Bacillus subtilis (natto) produces a capsular polypeptide of glutamate with a γ-linkage (called poly-γ-glutamate), which appears to serve as a physical barrier to phage adsorption. One ORF of PM1 had similarity with a poly-γ-glutamate hydrolase, which is assumed to degrade the capsular barrier to allow phage progenies to infect encapsulated host cells. The genome analysis of PM1 revealed the characteristics of the phage that are consistent as Bacillus subtilis (natto)-infecting phage.
Similar content being viewed by others
References
Y. Inatsu, N. Nakamura, Y. Yuriko, T. Fushimi, L. Watanasiritum, S. Kawamoto, Characterization of Bacillus subtilis strains in Thua nao, a traditional fermented soybean food in northern Thailand. Lett. Appl. Microbiol. 43, 237–242 (2006)
K. Jeyaram, W. Mohendro Singh, T. Premarani, A.R. Devi, K.S. Chanu, N.C. Talukdar, M.R. Singh, Molecular identification of dominant microflora associated with ‘Hawaijar’–a traditional fermented soybean (Glycine max (L.)) food of Manipur, India. Int. J. Food Microbiol. 122, 259–268 (2008)
D.Y. Kwon, J.W. Daily III, H.J. Kim, S. Park, Antidiabetic effects of fermented soybean products on type 2 diabetes. Nutr. Res. 30, 1–13 (2010)
P. Phromraksa, H. Nagano, T. Boonmars, C. Kamboonruang, Identification of proteolytic bacteria from Thai traditional fermented foods and their allergenic reducing potentials. J. Food Sci. 73, M189–M195 (2008)
N.N. Terlabie, E. Sakyi-Dawson, W.K. Amoa-Awua, The comparative ability of four isolates of Bacillus subtilis to ferment soybeans into dawadawa. Int. J. Food Microbiol. 106, 145–152 (2006)
H.A. Hong, J.-M. Huang, R. Khaneja, L.V. Hiep, M.C. Urdaci, S.M. Cutting, The safety of Bacillus subtilis and Bacillus indicus as food probiotics. J. Appl. Microbiol. 105, 510–520 (2008)
M. Schallmey, A. Singh, O.P. Ward, Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50, 1–17 (2004)
K. Iwai, N. Nakaya, Y. Kawasaki, H. Matsue, Inhibitory effect of natto, a kind of fermented soybeans, on LDL oxidation in vitro. J. Agric. Food Chem. 50, 3592–3596 (2002)
C.-L. Wang, T.B. Ng, F. Yuan, Z.K. Liu, F. Liu, Induction of apoptosis in human leukemia K562 cells by cyclic lipopeptide from Bacillus subtilis natto T-2. Peptides 28, 1344–1350 (2007)
Y. Kubo, A.P. Rooney, Y. Tsukakoshi, R. Nakagawa, H. Hasegawa, K. Kimura, Phylogenetic analysis of Bacillus subtilis strains applicable to natto (fermented soybean) production. Appl. Environ. Microbiol. 77, 6463–6469 (2011)
Y. Nishito, Y. Osana, T. Hachiya, K. Popendorf, A. Toyoda, A. Fujiyama, M. Itaya, Y. Sakakibara, Whole genome assembly of a natto production strain Bacillus subtilis natto from very short read data. BMC Genomics 11, 243 (2010)
D. Qui, K. Fujita, Y. Sakuma, T. Tanaka, Y. Ohashi, H. Ohshima, M. Tomita, M. Itaya, Comparative analysis of physical maps of four Bacillus subtilis (natto) genomes. Appl. Environ. Microbiol. 70, 6247–6256 (2004)
M. Itaya, K. Matsui, Conversion of Bacillus subtilis 168: natto producing Bacillus subtilis with mosaic genomes Biosci. Biotechnol. Biochem. 63, 2034–2037 (1999)
H. Brüssow, E. Kutter, in Bacteriophages: biology and applications, ed. by E. Kutter, A. Sulakvelidze (CRC Press, Boca Raton, 2005), pp. 129–163
K. Umene, S. Oohashi, F. Yamanaka, A. Shiraishi, Molecular characterization of the genome of Bacillus subtilis (natto) bacteriophage PM1. a phage associated with disruption of food production. World J. Microbiol. Biotechnol. 25, 1877–1881 (2009)
G. Bogosian, in The bacteriophages, ed. by R. Calendar (Oxford University Press, Oxford, 2006), pp. 667–673
M. Hongo, A. Yoshimoto, Bacteriophages of Bacillus natto part III. action of phage-induced γ-polyglutamic acid depolymerase on γ-polyglutamic acid and the enzymatic hydrolyzates. Agric. Biol. Chem. 34, 1055–1063 (1970)
K. Kimura, Y. Itoh, Characterization of poly-γ-glutamate hydrolase encoded by a bacteriophage genome: possible role in phage infection of bacillus subtilis encapsulated with poly-γ-glutamate. Appl. Environ. Microbiol. 69, 2491–2497 (2003)
S. Moineau, C. Lévesque, in Bacteriophages: biology and applications, ed. by E. Kutter, A. Sulakvelidze (CRC Press, Boca Raton, 2005), pp. 285–296
H.-W. Ackermann, in Bacteriophages: biology and applications, ed. by E. Kutter, A. Sulakvelidze (CRC Press, Boca Raton, 2005), pp. 67–89
E.J. Kim, J.W. Hong, N.-R. Yun, Y.N. Lee, Characterization of Bacillus phage-K2 isolated from chungkookjang, fermented soybean foodstuff. J. Ind. Microbiol. Biotechnol. 38, 39–42 (2011)
T. Nagai, F. Yamasaki, Bacillus subtilis (natto) bacteriophages isolated in Japan. Food Sci. Technol. Res. 15, 293–298 (2009)
J. Sambrook, D.W. Russell, Molecular cloning: a laboratory manual (Cold Spring Harbor Laboratory Press, New York, 2001)
T. Akimkina, C. Venien-Bryan, J. Hodgkin, Isolation, characterization and complete nucleotide sequence of a novel temperate bacteriophage Min1, isolated from the nematode pathogen Microbacterium nematophilum. Res. Microbiol. 158, 582–590 (2007)
A. Fujiwara, T. Kawasaki, S. Usami, M. Fujie, T. Yamada, Genomic characterization of Ralstonia solanacearum phage fRSA1 and its related prophage (fRSA1) in strain GMI1000. J. Bacteriol. 190, 143–156 (2008)
T. Yamada, S. Satoh, H. Ishikawa, A. Fujiwara, T. Kawasaki, M. Fujie, H. Ogata, A jumbo phage infecting the phytopathogen Ralstonia solanacearum defines a new lineage of the Myoviridae family. Virology 398, 135–147 (2010)
S.L. Salzberg, A.L. Delcher, S. Kasif, O. White, Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26, 544–548 (1998)
J. Xiong, Essential Bioinformatics (Cambridge University Press, New York, 2006)
T.M.A. Santos, R.C. Bicalho, Complete genome sequence of vB_EcoM_ECO1230-10: a coliphage with therapeutic potential for bovine metritis. Vet. Microbiol. 148, 267–275 (2011)
S.F. Altschul, W. Gish, W. Miller, E.W. Myers, D.J. Lipman, Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
R.H. Stevens, M.R. Ektefaie, D.E. Fouts, The annotated complete DNA sequence of Enterococcus faecalis bacteriophage fEf11 and its comparison with all available phage and predicted prophage genomes. FEMS Microbiol. Lett. 317, 9–26 (2011)
G. Bukovska, L. Klucar, C. Vlcek, J. Adamovic, J. Turna, J. Timko, Complete nucleotide sequence and genome analysis of bacteriophage BFK20-a lytic phage of the industrial producer Brevibacterium flavum. Virology 348, 57–71 (2006)
Z. Lu, E. Altermann, F. Breidt, P. Predki, H.P. Fleming, T.R. Klaenhammer, Sequence analysis of the Lactobacillus plantarum bacteriophage ΦJL-1. Gene 348, 45–54 (2005)
N. Jamalludeen, A.M. Kropinski, R.P. Johnson, E. Lingohr, J. Harel, C.L. Gyles, Complete genomic sequence of bacteriophage φEcoM-GJ1, a novel phage that has myovirus morphology and a podovirius-like RNA polymerase. Appl. Environ. Microbiol. 74, 516–525 (2008)
M.J. Mayer, J. Payne, M.J. Gasson, A. Narbad, Genomic sequence and characterization of the virulent bacteriophage φCTP1 from Clostridium tyrobutyricum and heterologous expression of its endolysin. Appl. Environ. Microbiol. 76, 5415–5422 (2010)
J.C. Alonso, G. Lüder, A.C. Stiege, S. Chai, F. Weise, T.A. Trautner, The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene 204, 201–212 (1997)
V. Lazarevic, A. Düsterhöft, B. Soldo, H. Hilbert, C. Mauël, D. Karamata, Nucleotide sequence of the Bacillus subtilis temperate bacteriophage SPβc2. Microbiology 145, 1055–1067 (1999)
A.D. Baxevanis, in Bioinformatics, ed. by A.D. Baxevanis, B.F.F. Ouellette (John Wiley & Sons, Inc., Hoboken, 2005), pp. 295–324
J.-M. Claverie, C. Notredame, Bioinformatics for dummies (Wiley Publishing, Inc., Indianapolis, 2007)
H. Brüssow, R.W. Hendrix, Phage genomics: small is beautiful. Cell 108, 13–16 (2002)
B. Guttman, R. Raya, E. Kutter, in Bacteriophages: biology and applications, ed. by E. Kutter, A. Sulakvelidze (CRC Press, Boca Raton, 2005), pp. 29–66
E. Kutter, R. Raya, K. Carlson, in Bacteriophages: biology and applications, ed. by E. Kutter, A. Sulakvelidze (CRC Press, Boca Raton, 2005), pp. 165–222
S. Petrovski, R.J. Seviour, D. Tillett, Genome sequence and characterization of the Tsukamurella bacteriophage TPA2. Appl. Environ. Microbiol. 77, 1389–1398 (2011)
A.V. Mardanov, N.V. Ravin, The antirepressor needed for induction of linear plasmid-prophage N15 belongs to the SOS regulon. J. Bacteriol. 189, 6333–6338 (2007)
A. Skowyra, S.A. MacNeill, Identification of essential and non-essential single-stranded DNA-binding proteins in a model archaeal organism. Nucleic Acids Res. 40, 1077–1090 (2012)
T.A. Baker, S.P. Bell, Polymerases and the replisome: machines within machines. Cell 92, 295–305 (1998)
M. Makowska-Grzyska, J.M. Kaguni, Primase directs the release of DnaC from DnaB. Mol. Cell 37, 90–101 (2010)
J.M. Kaguni, DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 60, 351–371 (2006)
W.K. Smits, H. Merrikh, C.Y. Bonilla, A.D. Grossman, Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J. Bacteriol. 193, 640–648 (2011)
C.E. Bell, Structure and mechanism of Escherichia coli RecA ATPase. Mol. Microbiol. 58, 358–366 (2005)
V.R.M. Chavali, C. Madhurantakam, S. Ghorai, S. Roy, A.K. Das, A.K. Ghosh, Genome segment 6 of Antheraea mylitta cypovirus encodes a structural protein with ATPase activity. Virology 377, 7–18 (2008)
A.-C. Déclais, D.M.J. Lilley, New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr. Opin. Struct. Biol. 18, 86–95 (2008)
I. Grainge, C. Lesterlin, D.J. Sherratt, Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 39, 5140–5148 (2011)
C.E. Catalano, in Viral genome packaging machines: genetics, structure, and mechanism, ed. by C.E. Catalano (Landes Bioscience/Eurekah.com, Georgetown, 2005), pp. 1–4
F. Arisaka, Assembly and infection process of bacteriophage T4. Chaos 15, 047502 (2005)
P.G. Leiman, S. Kanamaru, V.V. Mesyanzhinov, F. Arisaka, M.G. Rossmann, Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 60, 2356–2370 (2003)
B. Becker, N. de la Fuente, M. Gassel, D. Günther, P. Tavares, R. Lurz, T.A. Trautner, J.C. Alonso, Head morphogenesis genes of the Bacillus subtilis bacteriohage SPP1. J. Mol. Biol. 268, 822–839 (1997)
N.K. Abuladze, M. Gingery, J. Tsai, F.A. Eiserling, Tail length determination in bacteriophage T4. Virology 199, 301–310 (1994)
B.H. Yoon, H.I. Chang, Complete genomic sequence of the Lactobacillus temperate phage LF1. Arch. Virol. 156, 1909–1912 (2011)
T.G. Bernhardt, I.-N. Wang, D.K. Struck, R. Young, Breaking free: “protein antibiotics” and phage lysis. Res. Microbiol. 153, 493–501 (2002)
V.A. Fischetti, Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11, 393–400 (2008)
R. Young, I.-N. Wang, in The bacteriophages, ed. by R. Calendar (Oxford University Press, Oxford, 2006), pp. 104–125
D. Daniels, J. Schroeder, W. Szybalski, F. Sanger, F. Blattner, in Lambda II, ed. by R.W. Hendrix, J.W. Roberts, F.W. Stahl, R.A. Weisberg (Cold Spring Harbor Laboratory, USA, 1983), pp. 469–517
K.V. Srividhya, S. Krishnaswamy, Subclassification and targeted characterization of prophage-encoded two-component cell lysis cassette. J. Biosci. 32, 979–990 (2007)
C. Osera, G. Amati, C. Calvio, A. Galizzi, SwrAA activates poly-γ-glutamate synthesis in addition to swarming in Bacillus subtilis. Microbiology 155, 2282–2287 (2009)
Acknowledgments
Part of this study was supported by grants from the Ministry of Education, Science, Technology, Sports and Culture of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Umene, K., Shiraishi, A. Complete nucleotide sequence of Bacillus subtilis (natto) bacteriophage PM1, a phage associated with disruption of food production. Virus Genes 46, 524–534 (2013). https://doi.org/10.1007/s11262-013-0876-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-013-0876-4