Abstract
A gene of the Epstein-Barr virus (EBV), BamHI-C fragment rightward reading frame 1 (BCRF1), codes viral interleukin-10 (vIL-10), which is a close homolog to human IL-10. EBV strain variations are known at EBV latent membrane protein 1 (LMP1), and the distinct forms of LMP1 have been identified. In order to further elucidate the variations of EBV strains, the BCRF1 (vIL-10) gene was analyzed using PCR-direct sequencing in African Burkitt’s lymphoma (BL) cell lines Raji, P3HR-1, EB1 and Daudi, Japanese BL cell line Akata, lymphoblastoid cell line OB and 22 wild EBV isolates from eight gastric carcinoma tissues and 14 throat washes. We found only five variations of the vIL-10 gene in them with one silent mutation and three non-silent mutations. Raji had no mutation to the prototype gene of B95-8. EB1 and P3HR-1 had non-silent mutations in the sequences leading to the arginine/serine and threonine/proline interchanges at residues 4 and 166, respectively. The silent mutation was detected at valine 102 in Daudi and also in the Japanese cell lines Akata, OB and 20 (90.9%) of the wild EBV isolates. The type of variations in the vIL-10 gene had a common relationship with those in the LMP1 gene. All of the variants of valine 102 had China1-type LMP1 sequences except for Daudi with Med-type LMP1 and other minorities with B95-8 type LMP1. The conservativeness of vIL-10 with a few variations suggests the indispensability of the vIL-10 gene in EBV and that the variations of the vIL-10 gene may depend upon the geographical prevalence of the EBV strains. This is the first report regarding the variations of the vIL-10 gene in cell lines and other wild isolates.
Similar content being viewed by others
Introduction
Epstein-Barr virus (EBV) establishes an asymptomatic lifelong latent infection in the resting memory B cells in 95% of immunocompetent adults [1]. A gene of EBV, BamHI-C fragment rightward reading frame 1 (BCRF1) codes viral interleukin-10 (vIL-10) which is a close homolog to human IL-10 (hIL-10), with 84% amino acid homology [2]. The vIL-10 promoter is highly methylated and has no protein binding in Burkitt’s lymphoma (BL) cell lines or lymphoblastoid cell lines (LCLs) in agreement with an inactive vIL-10 promoter in the latently infected cells [3]. The vIL-10 gene is expressed only late in the lytic phase [4–7] and also early in EBV infection to human B cells [8], while hIL-10 is induced early in the lytic phase [4, 9]. hIL-10 and vIL-10 have B-cell growth factor activity [10, 11]. EBV-infected LCLs express hIL-10 and vIL-10 and promoted lymphoproliferative disease in immunocompromised mice [12]. hIL-10 and vIL-10 inhibit interferon (IFN)-gamma production by Th1 cells [2, 13, 14]. The LCLs infected by EBV recombinant with a mutant vIL-10 lacking C-terminal 54 amino acids important for normal vIL-10 function were less active or inactive in blocking IFN-gamma synthesis than the wild type vIL-10 recombinant EBV-infected LCLs [15]. The model mice infected with a recombinant vaccinia virus expressing IL-10 showed lower natural killer (NK) and CD8+ cytotoxic T lymphocyte (CTL) responses, suggesting that hIL-10 and vIL-10 may blunt the early NK cell and CTL responses so that EBV establishes a latent infection in the B-cell lymphocytes [16]. In addition, hIL-10 and vIL-10 downregulate class II major histocompatibility complex expression in monocytes/macrophages, thus resulting in the strong suppression of the proliferation of the helper T cells specific to the EBV-infected cells [17]. vIL-10 or hIL-10 induces the expression of EBV-encoded latent membrane protein 1 (LMP1) in B or NK cells infected by EBV with type II latency [18]. LMP1 mimics constitutively active CD40, induces the activation of NF-κB and the expressions of many genes, including hIL-10 [19]. In Hodgkin’s disease, LMP1 expression is induced by cytokines, such as IL-10 [20].
EBV strain variations are known at LMP1, EBV nuclear antigen 1 (EBNA1), EBNA2, EBNA3, and glycoprotein gp350/220 [21]. Seven phylogenetically distinct forms of LMP1 (B95-8, China1, China2, China3, Med, NC, AL) have been identified by their sequence differences and signature amino acid changes [22]. The LMP1 sequences of the Japanese EBV isolates are closely related to the China1 strain, which is a group of isolates found in Southern China [23, 24].
The sequence variation of the vIL-10 gene must be an interesting issue for virology, immunology, and oncology of EBV, but it has not yet been studied. In order to elucidate the variations of the vIL-10 gene in EBV strains, we examined the nucleotide sequence in EBV-positive cell lines and wild EBV isolates. The whole coding region of vIL-10 was characterized, and then the variations were compared with the LMP1 sequence variation. We found that the vIL-10 gene is stably conserved without or with a few variations in the tested samples. Such sequence variations of vIL-10 in the Japanese cell lines and wild isolates suggest the geographical distribution of the EBV strains. The conservativeness of vIL-10 gene suggests the importance of the vIL-10 function for virus evasion strategies.
Materials and methods
Cells, samples, and preparation of DNA
This work was approved by the Medical Ethics Committee for Human Subject Research at the Faculty of Medicine in Tottori University. Seven EBV-infected cell lines, eight EBV-positive gastric carcinoma (GC) tissues, and 14 EBV-positive throat washes (TW) were used in this study. The cell lines were cultured in a RPMI1640 medium (Nissui Pharmaceutical Co., Tokyo, Japan) with penicillin (100 IU/ml), streptomycin (100 μg/ml), and 10% fetal bovine serum in a CO2 incubator at 37°C. The tumor tissues of the GCs from GC patients were dissected from the stomachs removed at surgery. The EBV-positive GC samples were tested as previously described [23]. For the tissues and cells, DNA was extracted using a previously described method [23]. TW DNA was prepared by the alkaline lysis method as follows. Healthy volunteers gargled with 10 ml of phosphate-buffered saline for TW. The TWs were dehydrated by thrice repeated extractions with 10 ml 2-butanol, washed once with a 75% ethanol-25% H2O mixture, dissolved with 50 mM NaOH 300 μl, boiled for 10 min and neutralized by adding 1 M Tris–HCl, pH 7.5 to bring 138 mM Tris–HCl, pH 7.5. We tested 70 TWs for EBV genome-positiveness, based on PCR using a BamHI-W specific primer pair and found the positiveness in the 14 samples to be used in this study.
Direct sequencing of PCR products
PCR and direct sequencing were performed as described previously [23]. The method was modified to determine the sequence of the BCRF1 region as follows. Ten micro liters of PCR mixture were composed of 5 ng of the DNA sample, 1 U of Ex Taq polymerase (Takara Bio Inc., Otsu, Japan), supplemented Ex taq buffer, 0.25 mM dNTP, 0.5 μM sense primer 5′-gctctataaatcacttccctatctcaggta-3′ (Yama F; gene number 9627–9656) and antisense primer 5′-ggggtgaaatgcacccatctcctgcttcca-3′ (Yama R; 10230–10201), which were designed for up-stream and down-stream of the 513-bp coding region of the vIL-10 gene (gene number 9675–10187). PCR was performed in one cycle for 5 min at 95°C, 30 s at 55°C, and 1 min at 72°C, and in 24–34 cycles for 30 s at 95, 55°C, and 1 min at 72°C, respectively, using the PCR Thermal cycler MP TP3000 (Takara). The PCR products were subjected to electrophoresis on 1.8% agarose gel, and purified with S-400HR column (GE Healthcare, Piscataway, NJ). The cycle sequence reaction was performed as in the previous study [23]. The sequence data were checked for any homology in BLAST (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/) and any alignments between these data were analyzed using the Genetyx-SV RC software program, version 7.1 (Genetyx Co., Tokyo, Japan).
Phylogenetic analysis
In order to analyze the phylogenetic relationship of the vIL-10 and LMP1 genes, the vIL-10 sequences used the full length of their coding region (9675–10187), and LMP1 used the C-terminal of its coding region (168132–168594). The sequence data were aligned and the distances between each sample were calculated with the Clustal W (DNA Data Bank of Japan; http://www.ddbj.nig.ac.jp/search/clustalwj.html), using the Jukes-Cantor and Kimura two-parameter model. Phylogenetic trees were constructed from these matrices by the neighbor-joining method [25] using the Phylogenetic Tree Printer (Phylodendron; http://iubio.Bio.indiana.edu/treeapp/) from the PHYLIP formatted files.
Results
Viral interleukin-10 genes in EBV-positive cell lines
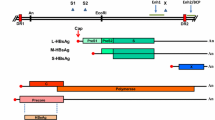
We examined the vIL-10 nucleotide sequence in the EBV-positive cell lines B95–8 [26], Raji [27], P3HR-1 [28], EB1 [29], Daudi [30], Akata [31], and OB [32] (Fig. 1). The EBV genome in the B95-8 cell line was used as the prototype EBV strain and the sequence of the vIL-10 gene was confirmed to be the same as that of the database (GenBank acc. No. V01555). Raji, P3HR-1, EB1, and Daudi were African BL cell lines, and Akata, and OB respectively were Japanese BL cell line and LCL. The Raji had the same sequence of vIL-10 as the B95-8 strain EBV. The P3HR-1 strain had a point mutation of the vIL-10 gene, an adenine (a)/cytosine (c) interchange at 10170 (a10170c) that corresponds to an amino acid change of threonine (T) at 166 into proline (P) (T166P). The EB1 strain had a guanine (g)/thymidine (t) interchange at gene number 9686 (g9686 t), thus resulting in the substitution of the arginine (R)/serine (S) interchange at residue 4 (R4S). The Daudi, Akata, and OB strains had a cytosine (c)/adenine (a) interchange at 9980 (c9980a), but the corresponding amino acid valine (V) at 102 (V102) was not changed. hIL-10 and vIL-10 are made of six helices (A-F) and five loops between the helices, and their receptor-binding sites are composed of helices A and F and the loop between helix A and helix B (AB loop)[33, 34]. The mutations of the vIL-10 gene were mapped on the amino acid sequence of the vIL-10 protein (Fig. 2).
Sequence variations of the vIL-10 gene and C-terminal of the LMP1 gene. The column across the top indicates the vIL-10 and LMP1 genes. The numbers across the second line correspond to the amino acid positions of the genes and the numbers across the third line correspond to the nucleotide positions under which the B95–8 prototype nucleotide sequence is listed. The names in the left column refer to the individual isolates. Only the sequences different from B95-8 are indicated. The small letters denote the nucleotides, and the amino acids are denoted by capital letters. An asterisk indicates a deletion of either a nucleotide or an amino acid
Amino acid sequence of the vIL-10 protein and the mutated amino acid on EBV-positive cell lines. The vIL-10 amino acid sequence of B95-8 is shown in capital letters. The amino acid numbers are shown in the upper lane. A signal peptide is shown as a black quadrangle, helix structures from A to F are shown in gray quadrangles and the locations of the mutated sequences (*) of cell lines are shown in parentheses below
Viral interleukin-10 genes in EBV isolates from TW and GC samples
We examined the vIL-10 sequences of the DNA samples from 14 TWs and eight EBV-positive Japanese GC tissues (Fig. 1). Of the 22 isolates, 19 isolates (86.4%) had only a point mutation of c9980a, which was detected in the Daudi, Akata, and OB strains. An isolate of TW8 had not only the mutation but also an additional non-silent mutation of c9843a, which resulted in the change of leucine (L) at 57 into isoleucine (I) (L57I). The other two isolates, TW13 and GC8, had the same sequence to B95-8. Therefore, only three types of variations of the vIL-10 gene were found in the EBV of the Japanese samples.
Latent membrane protein 1 sequence variations versus vIL-10 variations on cell lines, TW-isolates, and GC tissues
The sequence variations of LMP1 are classified into seven strains (B95-8, China1, China2, China3, Med, NC, AL)[22]. B95-8, Raji, EB1, and P3HR-1 are classified into the B95-8 strain and Daudi is classified into the Med (Mediterranean) strain. Previously, we determined the LMP1 nucleotide sequence of the EBVs in the cell lines [23]. Akata and OB are classified into the China1 strain. The China1 strain has characteristic mutations of c168357a, c168355t, the 30 bp-deletion and others in LMP1 gene [22]. We examined the LMP1 sequences of the Japanese 14 TWs and 8 GC tissues (Fig. 1). Of the total 22 EBV isolates, 18 isolates (82%) except for TW3, TW5, TW6, and TW13 had the similar variations to Akata or OB and were classified into the China1 strain in agreement with the classification [22]. TW3 and TW5 were classified into the China2 and B95-8 strains, respectively, and TW6 and TW13 were classified into the Med strain.
We characterized the variations of vIL-10 sequences with the variation of LMP1 sequences in the EBV strains and wild isolates and summarized the results (Table 1). B95-8, Raji, P3HR-1, and EB1 are classified into B95-8 strain by the LMP1 sequence [22]. They were classified into the same group of vIL-10 by the common sequence of c9980c. Although B95-8 and Raji had an identical vIL-10 sequence, but P3HR-1 and EB1 had respective non-silent mutations in vIL-10.
On the other hand, Daudi is classified in the Med strain by the LMP1 and had a silent mutation with c9980a of vIL-10. The c9980a of vIL-10 was also detected in 19 isolates (79%) with China1-type LMP1 of the 24 Japanese isolates except for TW3, TW5, TW6, TW13, and GC8. TW3, TW5, and TW6 had respectively China2-type LMP1, B95-8-type LMP1, and Med-type LMP1. TW13 had c9980c vIL-10 and had Med-type LMP1. GC8 had China1-type LMP1 with c9980c vIL-10. TW8 had China1-type LMP1 but had a unique non-silent mutation of vIL-10. Thus variations with c9980a of the vIL-10 sequences were mostly detected in EBVs with China1-type LMP1. This leads possibility that the variations of the vIL-10 gene suggest the geographical distribution of the EBV strains as well as LMP1.
In order to assign the Japanese EBV isolates, phylogenetic trees were drawn for the vIL-10 and LMP1 genes using nucleotide sequences (Fig. 3). The vIL-10 nucleotide sequences were divided into only five branches (Fig. 3A). Daudi, Akata, OB, and most of the Japanese isolates (86.4%) belonged into the same group. The other two isolates, TW13 and GC8, belonged to the B95-8 and Raji group, and TW8 formed an independent branch with the unique non-silent mutation. The LMP1 nucleotide sequences formed a node of a China1 with various variations between the Japanese specimens (Fig. 3B). The other specimens, TW6 and TW13, belonged to the same node as Daudi, and TW5 belonged to the node of B95-8. TW3 formed a branch, China 2.
Phylogenetic trees of vIL-10 and C-terminal of the LMP1 genes. Phylogenetic trees were drawn from the nucleotide sequence of vIL-10 (A) and the C-terminal of LMP1 (B). The isolates (TW: throat wash; GC: gastric carcinoma) are characteristically grouped according to their sequence variations. The scale shown in the lower right shows the evolutionary distance. This phylogenetic tree of LMP1 had little difference by the differences of the used isolates and the sequence domains to our previous report [23]
Discussion
This study demonstrates that the vIL-10 gene is stably conserved with a small difference in the nucleotide and amino acid sequences in different EBVs in cell lines and wild isolates. The mutations in the cell lines are shown in the amino acid sequence of the vIL-10 protein (Fig. 2). P3HR-1 and EB1 had an amino acid interchange. The T166P in P3HR-1 is interesting because the C-terminal sequence of hIL-10 (residues at 163–170, AYMTIKAR) represents the immunoinhibitory functions of hIL-10 [35]. The R4S substitution of vIL-10 in EB1 may evade such CTL responses. The vIL-10-producing lytic-cycle cells can be recognized by CTL because certain CTL clones targeted the vIL-10 peptide (residue 3-11) RRLVVTLQC in the context of HLA-B27 [36]. The functions of the mutated vIL-10 genes remain to be studied.
The sequence variation of vIL-10 found in the Daudi, Akata and OB and the majority of Japanese isolates had a silent mutation that codes for the same amino acid (V) (Fig. 1). This mutation will be useful as a marker for virus typing. The fact that Daudi, Akata, OB and the majority of Japanese isolates have the same mutation of vIL-10 could indicate that the EBV strains may stem from a common ancestral virus different from the other vIL-10 groups. Our data defined ten EBV variants in the Japanese EBVs by the typing of the vIL-10 and LMP1 genes as follows: Daudi(c9980a) -type vIL-10 and China1-type LMP1, China2-type LMP1, Med-type LMP1 or B95-8-type LMP1; B95-8-type vIL-10 and B95-8-type LMP1, Med-type LMP1 or China1-type LMP1; P3HR-1-type vIL-10 and B95-8-type LMP1; EB1-type vIL-10 and B95-8-type LMP1; TW8-type vIL-10 and China1-type LMP1. The c9980a-China1-type occupied 64% (9/14) of the TW isolates and 87% (7/8) of the GC isolates. This result suggests that the EBVs in Japanese are mixed populations indicated by variations of LMP1 and vIL-10 genes in EBVs from China, Mediterranean, and Africa, and the majority of EBV prevalence in Japan may be c9980a-China1.
Epstein-Barr virus has a long history of co-evolution with its host species. EBV isolates from Africa, European and Asia populations are distinguishable at polymorphic sites in the genome, reflecting the long period over which these host populations have been geographically isolated. These lie within the EBNA1, EBNA2, EBNA3, EBNA-LP, LMP1, and LMP2 genes as well as at other sites. The EBV-1 and EBV-2 (originally called A and B) have been identified on their EBNA2 and EBNA3 genes [21]. European populations carry EBV-1 strain mostly; however, EBV-2 strains appear to be almost equal prevalence with EBV-1 in part of equatorial Africa and in New Guinea.
We described that the Japanese EBV strains were different from the African EBVs such as Raji, P3HR-1, and the EB1 cell lines by the analysis of the LMP1 gene [23]. In this study, we report that the geographic and racial differences of the EBV isolates were seen in not only the LMP1 gene but also the vIL-10 gene. One can notice that Daudi is an exceptional African Burkitt’s lymphoma cell line with the c9980a-vIL-10 sequence. This may be explained by the fact that Daudi is classified into the Med strain, which is different from the major African strains B95-8, Raji, P3HR-1, and EB1 [22, 23]. We speculate that the majority of Japanese may have the c9980a-China1 strain which evolve via the Med strain rather than the African strains because Daudi is located near the phylogenetic branch of LMP1 to the Japanese isolates (Fig. 3B).
The sequence variations of vIL-10 were much smaller than those of LMP1, suggesting a weaker positive selection pressure for the IL-10 versus the LMP1 molecules [37, 38]. vIL-10 functions as a cytokine, similar to hIL-10 [14, 15, 18, 19]. It is thought that a small variation in the vIL-10 gene region enables it to evade host immunity because the vIL-10 protein mimics hIL-10. On the other hand, the LMP1 protein is a viral protein located on the cell membrane and is always exposed to host immunity. There was nothing the same nucleotide sequence of the LMP1 in the isolates (Fig. 1). It is thought that LMP1 evades host immunity by having multiple variations as a result of positive selection [37]. These results suggest that EBV can evolve under the exposure to host immunity and survive in various substrains.
In conclusion, we identified a few point mutations of the vIL-10 gene in cultured cell lines and wild isolates. The vIL-10 gene is conserved stably in EBV strains and wild isolates and will be useful as a marker for EBV typing.
References
A.B. Rickinson, E. Kieff, in Epstein-Barr virus, ed. by D.M. Knipe, P.M. Howley, D.E. Griffin (Lippincott Williams & Wilkins, Philadelphia, 2001), pp. 2575–2627
P. Vieira, R. de Waal-Malefyt, M.N. Dang, K.E. Johnson, R. Kastelein, D.F. Fiorentino, J.E. deVries, M.G. Roncarolo, T.R. Mosmann, K.W. Moore, Proc. Natl. Acad. Sci. U S A 88, 1172–1176 (1991)
H.H. Niller, D. Salamon, M. Takacs, J. Uhlig, H. Wolf, J. Minarovits, Biol. Chem. 382, 1411–1419 (2001)
T. Sairenji, E. Ohnishi, S. Inouye, T. Kurata, Viral. Immunol. 11, 221–231 (1998)
R. Touitou, C. Cochet, I. Joab, J. Gen. Virol. 77(Pt 6), 1163–1168 (1996)
J.P. Stewart, F.G. Behm, J.R. Arrand, C.M. Rooney, Virology 200, 724–732 (1994)
G.S. Hudson, A.T. Bankier, S.C. Satchwell, B.G. Barrell, Virology 147, 81–98 (1985)
I. Miyazaki, R.K. Cheung, H.M. Dosch, J. Exp. Med. 178, 439–47 (1993)
S. Mahot, A. Sergeant, E. Drouet, H. Gruffat, J. Gen. Virol. 84, 965–974 (2003)
F. Rousset, E. Garcia, T. Defrance, C. Peronne, N. Vezzio, D.H. Hsu, R. Kastelein, K.W. Moore, J. Banchereau, Proc. Natl. Acad. Sci. U S A 89, 1890–1893 (1992)
A.D. Stuart, J.P. Stewart, J.R. Arrand, M. Mackett, Oncogene 11, 1711–1719 (1995)
G.K. Hong, M.L. Gulley, W.H. Feng, H.J. Delecluse, E. Holley-Guthrie, S.C. Kenney, J. Virol. 79, 13993–14003 (2005)
K.W. Moore, P. Vieira, D.F. Fiorentino, M.L. Trounstine, T.A. Khan, T.R. Mosmann, Science 248, 1230–1234 (1990)
D.H. Hsu, R. de Waal Malefyt, D.F. Fiorentino, M.N. Dang, P. Vieira, J. de Vries, H. Spits, T.R. Mosmann, K.W. Moore, Science 250, 830–832 (1990)
S. Swaminathan, R. Hesselton, J. Sullivan, E. Kieff, J. Virol. 67, 7406–7413 (1993)
M.G. Kurilla, S. Swaminathan, R.M. Welsh, E. Kieff, R.R. Brutkiewicz, J. Virol. 67, 7623–7628 (1993)
R. de Waal Malefyt, J. Haanen, H. Spits, M.G. Roncarolo, A. te Velde, C. Figdor, K. Johnson, R. Kastelein, H. Yssel, J.E. de Vries, J. Exp. Med. 174, 915–924 (1991)
L.L. Kis, M. Takahara, N. Nagy, G. Klein, E. Klein, Blood 107, 2928–2935 (2006)
H. Nakagomi, R. Dolcetti, M.T. Bejarano, P. Pisa, R. Kiessling, M.G. Masucci, Int. J. Cancer 57, 240–4 (1994)
L.L. Kis, M. Takahara, N. Nagy, G. Klein, E. Klein, Immunol. Lett. 104, 83–8 (2006)
E. Kieff, A.B. Rickinson, in Epstein-Barr virus, ed. by D.M. Knipe, P.M. Howley, D.E. Griffin (Lippincott Williams & Wilkins, Philadelphia, 2001), pp. 2511–2573
R.H. Edwards, F. Seillier-Moiseiwitsch, N. Raab-Traub, Virology 261, 79–95 (1999)
K. Kanai, Y. Satoh, Y. Saiki, H. Ohtani, T. Sairenji, Virus Genes 34, 55–61 (2007)
Y. Wang, K. Kanai, Y. Satoh, B. Luo, T. Sairenji, Intervirology 50, 229–236 (2007)
N. Saitou, M. Nei, Mol. Biol. Evol. 4, 406–425 (1987)
G. Miller, T. Shope, H. Lisco, D. Stitt, M. Lipman, Proc. Natl. Acad. Sci. U S A 69, 383–387 (1972)
J.V. Pulvertaft, J. Clin. Pathol. 18, 261–273 (1965)
Y. Hinuma, M. Konn, J. Yamaguchi, D.J. Wudarski, J.R. Blakeslee Jr., J.T. Grace Jr., J. Virol. 1, 1045–1051 (1967)
M.A. Epstein, Y.M. Barr, J. Natl. Cancer Inst. 34, 231–240 (1965)
E. Klein, G. Klein, J.S. Nadkarni, J.J. Nadkarni, H. Wigzell, P. Clifford, Lancet 2, 1068–1070 (1967)
K. Takada, Int. J. Cancer 33, 27–32 (1984)
E. Ohnishi, T. Iwata, S. Inouye, T. Kurata, T. Sairenji, J. Interferon Cytokine Res. 17, 597–602 (1997)
A. Zdanov, C. Schalk-Hihi, S. Menon, K.W. Moore, A. Wlodawer, J. Mol. Biol. 268, 460–467 (1997)
S.I. Yoon, B.C. Jones, N.J. Logsdon, M.R. Walter, Structure 13, 551–564 (2005)
B. Gesser, H. Leffers, T. Jinquan, C. Vestergaard, N. Kirstein, S. Sindet-Pedersen, S.L. Jensen, K. Thestrup-Pedersen, C.G. Larsen, Proc. Natl. Acad. Sci. U S A 94, 14620–14625 (1997)
X. Saulquin, M. Bodinier, M.A. Peyrat, A. Hislop, E. Scotet, F. Lang, M. Bonneville, E. Houssaint, Eur. J. Immunol. 31, 708–715 (2001)
J.M. Burrows, L. Bromham, M. Woolfit, G. Piganeau, J. Tellam, G. Connolly, N. Webb, L. Poulsen, L. Cooper, S.R. Burrows, D.J. Moss, S.M. Haryana, M. Ng, J.M. Nicholls, R. Khanna, J Virol. 78, 7131–7137 (2004)
Z. Yang, R. Nielsen, N. Goldman, A.M. Pedersen, Genetics 155, 431–449 (2000)
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (No. B13390150) and Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the 2nd Term Comprehensive 10-year Strategy for Cancer Control from Ministry of Health and Welfare of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kanai, K., Satoh, Y., Yamanaka, H. et al. The vIL-10 gene of the Epstein-Barr virus (EBV) is conserved in a stable manner except for a few point mutations in various EBV isolates. Virus Genes 35, 563–569 (2007). https://doi.org/10.1007/s11262-007-0153-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-007-0153-5