Abstract
Rice tungro is a viral disease seriously affecting rice production in South and Southeast Asia. Tungro is caused by the simultaneous infection in rice of Rice tungro bacilliform virus (RTBV), a double-stranded DNA virus and Rice tungro spherical virus (RTSV), a single-stranded RNA virus. To apply the concept of RNA-interference (RNAi) for the control of RTBV infection, transgenic rice plants expressing DNA encoding ORF IV of RTBV, both in sense as well as in anti-sense orientation, resulting in the formation of double-stranded (ds) RNA, were raised. RNA blot analysis of two representative lines indicated specific degradation of the transgene transcripts and the accumulation of small molecular weight RNA, a hallmark for RNA-interference. In the two transgenic lines expressing ds-RNA, different resistance responses were observed against RTBV. In one of the above lines (RTBV-O-Ds1), there was an initial rapid buildup of RTBV levels following inoculation, comparable to that of untransformed controls, followed by a sharp reduction, resulting in approximately 50-fold lower viral titers, whereas the untransformed controls maintained high levels of the virus till 40 days post-inoculation (dpi). In RTBV-O-Ds2, RTBV DNA levels gradually rose from an initial low to almost 60% levels of the control by 40 dpi. Line RTBV-O-Ds1 showed symptoms of tungro similar to the untransformed control lines, whereas line RTBV-O-Ds2 showed extremely mild symptoms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice tungro disease, the most important viral disease of rice, is widespread in South and Southeast Asia and is believed to be responsible for annual losses nearing 109 US dollars worldwide (Herdt 1991). More recent estimates reveal that the disease causes on an average about 2% losses in rice production in India, although at the regional level, losses can be more significant (Muralidharan et al. 2003). Rice tungro is caused by the joint infection of two unrelated viruses Rice tungro bacilliform virus (RTBV), a double-stranded DNA-containing virus, belonging to the genus Tungrovirus and Rice tungro spherical virus (RTSV), a single-stranded RNA virus belonging to the genus Waikavirus (Jones et al. 1991). RTBV and RTSV, also known as the “Tungro virus complex”, are transmitted exclusively by the Green leafhopper, GLH, Nephotettix virescens (Hibino and Cabauatan 1987). The most conspicuous symptoms of tungro are the stunting of plants and yellow-orange discolouration of leaves, both of which are believed to be caused by RTBV, as observed in symptomatic plants subjected to Agrobacterium-mediated inoculation of the virus (Dasgupta et al. 1991).
Because of its role in limiting rice production, incorporation of tungro resistance has been an important breeding objective in rice improvement programs in Asia. Several sources of genetic resistance have been reported in rice against RTSV, but against RTBV, there are only a few (Azzam and Chancellor 2002). Additionally, none of the host resistance sources have been genetically well-characterized. Thus, in order to ensure durability of the otherwise fragile resistance under field conditions (Manwan et al. 1985; Dahal et al. 1990), transgenic strategies for tungro resistance, targeting RTBV are promising.
Two strategies, namely protein-mediated and RNA-mediated have been the underlying principles behind most successful transgenic viral resistance. Both strategies have emanated from the concept of “pathogenderived resistance” or PDR (Sanford and Johnston 1985), wherein pathogen-encoded proteins or RNA are used to interfere with crucial steps in the infection cycle. More recently, RNA-mediated resistance has been shown to be robust and widely applicable against several classes of viral pathogens in plants (Mansoor et al. 2006).
Homology-dependent selective degradation of RNA, RNA-interference (RNAi) or Post-transcriptional gene silencing (PTGS) is involved in several biological phenomena, including adaptive defence against viruses in plants (Ratcliffe et al. 1999; Vance and Vaucheret 2001; Yu and Kumar 2003; Herr 2005). The first demonstration of RNAi-mediated virus resistance was shown by Waterhouse et al. (1998), against Potato virus Y (PVY) in transgenic tobacco plants. Resistance against PVY in transgenic tobacco plants expressing the PVY protease gene simultaneously in sense and anti-sense orientation was much higher than in lines expressing the same gene individually in either orientation. Subsequently, the same principle has been exploited successfully in different host systems to obtain resistance against several other viruses (Pooggin et al. 2003; Vanitharani et al. 2003; Tenllado et al. 2003; Di Nicola-Negri et al. 2005; Lennefors et al. 2006; Abhary et al. 2006).
Here we report on the use of RNAi to confer resistance against RTBV in rice. We show that transgenic rice plants expressing a RTBV gene in both sense as well as anti-sense orientation, showed only mild tungro symptoms following challenge inoculation with the virus, whereas plants expressing the same gene in sense orientation were found to be as symptomatic as untransformed control plants.
Materials and methods
Plasmid constructs
A DNA fragment between nucleotide residues 5700 and 7026 encoding the RTBV open reading frame IV (ORFIV) of an Indian isolate RTBV-AP was obtained by BglII digestion of the plasmid pRTBV204 (Nath et al. 2002). This fragment was cloned in between Cauliflower mosaic virus 35S promoter and nopaline synthase transcription termination signals separately in sense and anti-sense orientations. The entire cassette of the above promoter-ORFIV-terminator fragment was inserted at SmaI site of the binary plasmid pCAMBIA1380 (Roberts et al. 1998) to obtain plasmid pRTBV-Os (Fig. 1a). ORFIV in pRTBV-Os is untranslatable since it lacks the initiation codon. pRTBV-O-Ds (Fig. 1a) was derived from pRTBV-Os by cloning an additional copy of the promoter-ORFIV-terminator cassette, having the gene in anti-sense orientation in the end-filled BglII site of pRTBV-Os.
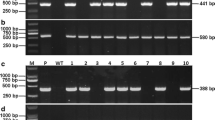
Analysis of transgenic plants to confirm their transgenic nature. Panel a shows schematic map of the T-DNA regions of the DNA plasmid constructs used to raise transgenic plants. The upper part shows pRTBV-Os and the lower part, pRTBV-O-Ds. P: CaMV 35S promoter, T: nopaline synthase transcription terminator, ORFIV: RTBV ORFIV, TL: Left border of T-DNA region, TR: Right border of T-DNA region. Panel b Southern hybridization of two transgenic lines generated using pRTBV-O-Ds. Lane 1: RTBV-O-Ds1, 2: RTBV-O-Ds2, C: untransformed control plant. The numbers at the side indicate the positions of size markers in kb. The restriction enzyme used was KpnI. The probe used was a DNA fragment encoding RTBV ORFIV
Generation of transgenic rice plants
The binary vectors (pRTBV-Os and pRTBV-O-Ds) were transferred into a virulent Agrobacterium tumefaciens strain EHA105 (Holsters et al. 1978). A culture obtained from a recombinant Agrobacterium colony was used to transform the rice variety Pusa Basmati-1 by the method of Wang et al. (1997).
RNA analysis
In order to ascertain transcript levels of the inserted transgene, northern analysis was performed using total RNA isolated by the method of Chomczynski and Sacchi (1987) from 20-day old transgenic plants. Fractions enriched with small RNA were obtained and transferred on to the membrane using the method of Hamilton and Baulcombe (1999). The ORFIV fragment was radiolabelled by employing Random Primer Labeling Kit (Roche) and α-32P-dATP (specific activity 3000 Ci/mmol, BRIT, India) as per manufacturer’s specifications. Hybridization and washes were carried out in 7% SDS, 50% deionized formamide at 42°C and twice in 2 × SSC, 0.2% SDS at 42°C for 30 min, respectively.
Viral resistance assay
RTBV and RTSV were maintained in rice plants under glasshouse conditions by GLH-mediated serial transfer, initiated using naturally-infected plants. GLH reared on healthy rice plants were enclosed with such plants for virus acquisition in a mylar cage for 24 h. A group of five 15–20 day-old T1 transgenic plants representing a single transformation event were inoculated together for 24 h with 3 viruliferous GLH per plant, obtained as described above. As a control group, five non-transgenic plants were also inoculated in a similar manner. Both groups were maintained at 30°C with 14/10 h photoperiod with supplementary lighting under greenhouse conditions to allow the build up of virus levels and development of symptoms. Approximately 1–2 cm samples were collected separately from the second leaf of each of the transgenic and non-transgenic plants at 10-day intervals for a period of 40 days. Samples collected from transgenic plants and those from non-transgenic plants were pooled separately for each day of sampling as a transgenic pool and a non-transgenic pool. The extent of viral DNA accumulation in the above two pools were then assessed by slot-blot hybridization of DNA isolated from the pooled samples. Similar hybridization and autoradiography conditions were maintained to achieve uniformity and reproducibility of results. The hybridization signals were then converted to numerical values by densitometric scanning, which were used as a measure of the viral DNA titer in each sample. The linear relationship of the hybridization signal and the numerical values obtained by densitometric scanning was checked by using known quantities of cloned viral DNA. The resistance assay was performed twice for all lines tested.
Results
Integrated transgenes present at low copy numbers in transgenic rice plants
A total of approximately 40 putative transgenic rice plants were obtained using pRTBV-Os and pRTBV-O-Ds. On Southern analysis of about 10 plants derived from pRTBV-O-Ds (Fig. 1a, lower panel), two classes of insertion patterns, showing different electrophoretic mobilities were obtained following KpnI digestion of total DNA, probed with ORFIV (two representative patterns shown in Fig. 1b). Since KpnI has no recognition site between the two T-DNA borders, the number of fragments hybridizing with the probe indicates the number of insertion sites of the transgene. Since tandem insertion of multiple copies of the transgene is rare, it was assumed that the above indicates that one single-copy and another double-copy transgenic event has been obtained, which were named RTBV-O-Ds1 (Fig. 1b, lane 1) and RTBV-O-Ds2 (Fig. 1b, lane 2) and represented integration of the same transgene at two different sites in the genome. Untransformed plant (Fig. 1b, lane c) showed no hybridization, indicating that the hybridization was specific to the transgene. A similar analysis of plants derived using pRTBV-Os (Fig. 1a, upper panel) showed a pattern suggestive of single copy integration event (data not shown).
Transgene-specific transcripts accumulated to low levels in RTBV-O-Ds plants but to much higher levels in RTBV-Os plants
On northern analysis, total RNA isolated from T1 progenies of both RTBV-O-Ds1 and RTBV-O-Ds2 revealed very low or undetectable levels of transcripts representing ORFIV. As shown in the upper panel of Fig. 2a, except the plant represented in lane 3, none of the plants accumulated detectable transcripts homologous to the probe (ORFIV). In contrast, transcripts of the selection marker hygromycin phosphotransferase (hpt) accumulated to high levels in all the lines tested (middle panel). The untransformed control plant did not accumulate transcripts homologous to the probe used (Fig. 2a, lane c). The loading control shown in the lower panel indicates equal loading of RNA in all lanes. On using the same probe, a similar analysis of 12 plants derived from RTBV-Os revealed low levels of transcript accumulation in three plants (Fig. 2b, lanes 1, 4 and 5) and high levels in the rest of the nine plants (lanes 2, 3 and 6–12), the control non-transgenic sample showing no hybridization (lane c).
Transcript analysis of transgenic plants to determine the accumulation of transgene-specific transcripts. Panel a northern blot analysis of total RNA extracted from T1 progenies of line RTBV-O-Ds1 (lanes 1 and 2), line RTBV-O-Ds2 (lanes 3–5) and untransformed plant (lane C), the upper panel being probed with RTBV ORFIV, the middle panel with hpt gene and the lower panel showing total RNA as the loading control. Panel b northern blot analysis of T1 progenies of line RTBV-Os (lanes 1–12) and control non-transgenic plant (lane C), the upper panel probed with ORFIV and the lower panel showing total RNA as the loading control. Panel c shows small RNA extracted from T1 progenies of lines RTBV-O-Ds2 (lanes 1–3), RTBV-O-Ds1 (lanes 4 and 5) and control untransformed plants (lane C), probed with fragmented ORFIV gene. The estimated sizes are indicated at the sides
Low-molecular-weight RNAs accumulated in RTBV-O-Ds1 and RTBV-O-Ds2 lines
When the accumulation of low molecular weight RNAs, indicative of specific RNA degradation in the transgenic plants was investigated, 22–23 bp species, specific to ORFIV were seen to accumulate in all the three RTBV-O-Ds2 lines (Fig. 2c, lanes 1–3) and one out of two RTBV-O-Ds1 lines tested (Fig. 2c, lane 5).
RTBV accumulation pattern differed in inoculated transgenic lines and controls
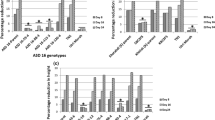
Following GLH-mediated inoculation, RTBV accumulated rapidly in non-transgenic control plants within 10 days post-inoculation (dpi) and remained high throughout the course of the experiment. Average RTBV titers of each pooled sample, expressed as a percent fraction of the corresponding value in the non-transgenic control pool are represented in Fig. 3. In RTBV-O-Ds1, RTBV accumulation was similar to that of the control sample at 10 dpi, but dropped to half the above levels by 20 dpi and became barely detectable by 30 dpi. In RTBV-O-Ds2 lines, RTBV accumulation was more gradual, increasing steadily to 60% of the control levels by the end of the experiment. In contrast, RTBV-Os lines exhibited a pattern of RTBV accumulation similar to that of the controls (data not shown).
Relative RTBV DNA titers at different days post-inoculation (dpi) in transgenic lines RTBV-O-Ds1, RTBV-O-Ds2 and non-transgenic control plants, following GLH-mediated inoculation of RTBV. The titers in the control plants at 10–40 dpi have been shown as 100% and the titers in the pooled samples of test plants shown as percent fraction. Each value is an average of two independent experiments
RTBV-O-Ds transgenic plants showed moderate to mild tungro symptoms
Following GLH-mediated inoculation, within 20 dpi, control untransformed plants developed typical tungro symptoms, namely stunting and yellowing of leaves. Using the same inoculation conditions, progenies of lines RTBV-Os and RTBV-O-Ds1 showed symptoms similar to the controls, whereas RTBV-O-Ds2 plants showed mild stunting. At 5 months post-inoculation, almost half of the RTBV-O-Ds2 plants grew normally and flowered (Fig. 4, T) as compared to non-transgenic control plants which were severely stunted and did not flower (Fig. 4, C).
Discussion
There are several reports on the use of RNAi to obtain virus resistance in plants (Vanitharani et al. 2003; Tenllado et al. 2003; Abhary et al. 2006; Lennefors et al. 2006; Ramesh et al. 2007). In one of the first such reports, a 65–68% reduction in the transient accumulation of the DNA of African cassava mosaic virus in tobacco protoplasts was reported (Vanitharani et al. 2003), when co-inoculated with siRNAs directed against the transcripts of replication-associated protein (Rep) of the virus. Similarly, Tenllado et al. (2003) showed a strong reduction in the titers of Pepper mild mottle virus and Plum pox virus in Nicotiana benthamiana, when the respective virus was co-inoculated or the plants sprayed with bacterially-expressed virus-specific siRNAs. Abhary et al. (2006) reported resistance against several geminiviruses associated with Tomato yellow leaf curl disease in tomato and tobacco, following agroinfiltration of the plants with RNAi-initiating constructs against conserved regions of the viral genomes. In a recent report (Chilakamurthi et al. 2007), it is shown that ribozyme-mediated degradation of Rep transcripts of the geminivirus Mungbean yellow mosaic India virus resulted in 65% decrease in geminiviral replication in a yeast model system. Similarly natural (Lennefors et al. 2006; Tougou et al. 2006; Bonfim et al. 2007; Ramesh et al. 2007) and experimental (Di Nicola-Negri et al. 2005) plant hosts, and in some cases, both (Batuman et al. 2006) have been transformed with hairpin RNA-expressing constructs derived from various viral genes, designed to initiate RNAi and which showed varying degrees of viral resistance.
Two earlier reports of transgenic resistance against RTSV, the viral component responsible for transmission of the tungro virus complex by GLH have been published (Sivamani et al. 1999; Huet et al. 1999). In this report, we have examined viral resistance in rice against an Indian strain of RTBV, RTBV-AP using double-stranded RNA-generating construct and compared it with that in plants containing the corresponding single-stranded RNA. RTBV produces a single full-length transcript to translate three out of four ORFs, the fourth (ORFIV) is translated by a spliced transcript consisting of ORFIV coding sequences directly joined to the 5’ untranslated region (Futterer et al. 1994). In addition, the full-length transcript also acts as the template for the reverse transcription-mediated replication of the viral DNA, a common step in all pararetroviruses (Hull 2002). Hence, by virtue of the dual functions performed by their transcripts, pararetroviruses appear to be especially suitable for RNAi-mediated inhibition of replication. Since no other report on the use of RNAi targeting pararetroviruses exists in the literature, decision on the suitability of a particular viral gene for successful initiation of RNAi was based on the importance of targeting all viral transcripts. Keeping in view the splicing mentioned above, the decision of utilizing ORFIV for the above purpose was to target all transcripts of RTBV, both spliced as well as unspliced. This would not have been possible with any other RTBV DNA fragment.
Compared to most reports of RNAi-mediated viral resistance in plants which generate hairpin-loop RNA, our strategy used double-stranded RNA without a hairpin-loop as template. The absence of transcript accumulation of the transgene in the Ds lines tested (Fig. 2a) coupled with the presence of corresponding low molecular weight RNA (22–23 bp) in both RTBV-O-Ds1 and RTBV-O-Ds2 lines (Fig. 2c) indicated that RNAi is active, illustrating the effectiveness of this strategy. On challenging the two Ds lines with viruliferous GLH, differing patterns of resistance response, as assessed by the accumulation of the viral DNA was seen, despite both lines carrying copies of the same transgene. It is possible that the two lines transcribed the transgene to different levels because of differing levels of promoter activity at two independent locations in the genome as a result of altered methylation levels as reported earlier (Meyer 1998), resulting in differing levels of RNAi.
Of the two lines tested, RTBV-O-Ds1 plants showed tungro symptoms similar to non-transgenic controls whereas RTBV-O-Ds2 plants showed symptoms of very mild stunting. Therefore, symptoms, believed to be caused by RTBV alone, may express due to differing temporal buildup of RTBV titers in the two lines. It is noteworthy that in RTBV-O-Ds1, RTBV titers were close to the high value in non-transgenic controls at 10 dpi itself, but rapidly declined and became almost undetectable by 30 dpi, whereas the titer increased gradually in RTBV-O-Ds2 (Fig. 3), remaining quite low in the period between 10–20 dpi. It is possible that the latter period (which is about 25–35 days post-germination) is crucial for proper growth in rice plants and high viral titers during that period have a profound and permanent effect on normal plant growth as was also observed by Santa Cruz et al. (2003). This might explain why RTBV-O-Ds2 plants showed mild symptoms, presumably because the viral titers are relatively low during the above period and gradually accumulate to a high level later on. It is possible that symptoms result from differential modulation of development-specific host micro-RNA levels in the above two lines triggered by the infecting RTBV through RNAi, as has been reported for Cauliflower mosaic virus, a pararetrovirus and in Arabidopsis thaliana (Moissard and Voinnet 2006).
In conclusion, RTBV-O-Ds2 line, by virtue of developing mild tungro symptoms on challenge with viruliferous GLH, represents an important step in the development of tungro resistance in rice suitable for Indian conditions. Analysis of two complete and one partial length RTBV genomic sequences from widely separated sites in India has already shown their highly conserved nature (Nath et al. 2002; Verma and Dasgupta 2007), indicating that such lines can be used as important breeding material for transferring the transgene to various genetic backgrounds of rice more suited for cultivation in various tungro-threatened rice growing regions of India.
References
Abhary MK, Anfoka GH, Nakhla MK, Maxwell DP (2006) Post-transcriptional gene silencing in controlling viruses of the Tomato yellow leaf curl virus complex. Arch Virol 151:2349–2363
Azzam O, Chancellor TCB (2002) The biology, epidemiology and management of Rice tungro disease in Asia. Plant Dis 86:88–100
Batuman O, Mawassi M, Bar-Joseph M (2006) Transgenes consisting of a dsRNA of an RNAi suppressor plus the 3′ UTR provide resistance to Citrus tristeza virus sequences in Nicotiana benthamiana but not in citrus. Virus Genes 33:319–327
Bonfim K, Faria JC, Nogueira EO, Mendes EA, Aragão FJ (2007) RNAi-mediated resistance to Bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol Plant Microbe Interact 20:717–726
Chilakamurthi U, Mukherjee SK, Deb JK (2007) Intervention of geminiviral replication in yeast by ribozyme mediated downregulation of its Rep protein. FEBS Lett 581:2675–2683
Chomczynski P, Sacchi N (1987) Single step method of RNA isolation by acid guanidium thiocyanate phenol chloroform extraction. Anal Biochem 162:156–159
Dahal G, Hibino H, Cabunagan RC, Tiongco ER, Flores ZM, Aguiero VM (1990) Changes in cultivar reaction to tungro due to changes in “virulence” of the leafhopper vector. Phytopathology 80:659–665
Dasgupta I, Hull R, Eastop S, Poggi-pollini C, Blakebrough M, Boulton MI, Davies JW (1991) Rice tungro bacilliform virus DNA independently infects rice after Agrobacterium-mediated transfer. J Gen Virol 72:1215–1221
Di Nicola-Negri E, Brunetti A, Tavazza M, Ilardi V (2005) Hairpin RNA-mediated silencing of Plum pox virus P1 and HC-Pro genes for efficient and predictable resistance to the virus. Transgenic Res 14:989–994
Futterer J, Potrykus I, Valles-Brau MP, Dasgupta I, Hull R, Hohn T (1994) Splicing in a plant pararetrovirus. Virology 198:663–670
Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952
Herdt RW (1991) Research priorities for biotechnology. In: Khush GS, Toennissen GH (eds) Rice biotechnology. CAB International, Wallingford, UK, pp 19–54
Herr AJ (2005) Pathways through the small RNA world of plants. FEBS (Fed Eur Biol Soc) Lett 579:5879–5888
Hibino H, Cabauatan PQ (1987) Infectivity neutralization of rice tungro-associated viruses acquired by vector leafhoppers. Phytopathology 77:473–476
Hibino H, Saleh N, Roechan M (1979) Transmission of two kinds of rice tungro-associated viruses by insect vectors. Phytopathology 69:1266–1268
Holsters M, Waele D, Depicker A, Messens E, Van Montagu M, Schell J (1978) Transfection and transformation of A. tumefaciens. Mol Gen Genet 168:181–187
Huet H, Mahendra S, Wang J, Sivamani E, Ong CA, Chen L, de Kochko A, Beachy RN, Fauquet C (1999) Near imunity to Rice tungro spherical virus achieved in rice by a replicase-mediated resistance strategy. Phytopathology 89:1022–1027
Hull RM (2002) Plant virology, 4th edn. Academic Press, London
Jones MC, Gough K, Dasgupta I, Subba Rao BL, Cliffe J, Qu R, Shen P, Kaniewska M, Blakebrough M, Davies JW, Beachy RN, Hull R (1991) Rice tungro disease is caused by an RNA and a DNA virus. J Gen Virol 72:757–761
Lennefors B-L, Savenkov EI, Bensefelt J, Wremerth-Weich E, van Roggen P, Tuvesson S, Valkonen JPT, Gielen J (2006) dsRNA-mediated resistance to Beet necrotic yellow vein virus infections in sugar beet (Beta vulgaris L. ssp. vulgaris). Mol Breeding 18:313–325
Mansoor S, Amin I, Hussain M, Zafar Y, Briddon RW (2006) Engineering novel traits in plants through RNA-interference. Trends Plant Sci 11:559–565
Manwan I, Sama S, Rizvi SA (1985) Use of varietal rotation in the management of tungro disease in Indonesia. Indones. Agric Res Dev J 7:43–48
Meyer P (1998) Stabilities and instabilities in transgene expression. In: Lindsey K (ed) Transgenic plant research. Harwood Academic Publishers, Amsterdam, The Netherlands, pp 263–275
Moissard G, Voinnet O (2006) RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA 103:19593–19598
Muralidharan K, Krishnaveni D, Rajarajeshwari NVL, Prasad ASR (2003) Tungro epidemic and yield losses in paddy fields in India. Curr Sci 85:1143–1147
Nath N, Mathur S, Dasgupta I (2002) Molecular analysis of two complete rice tungro bacilliform virus sequences from India. Arch Virol 147:1173–1187
Pooggin M, Sivaprasad PV, Veluthambi K, Hohn T (2003) RNAi targeting of DNA virus in plants. Nature Biotech 21:131–132
Ramesh SV, Mishra AK, Praveen S (2007) Hairpin RNA-mediated strategies for silencing of Tomato leaf curl virus AC1 and AC4 genes for effective resistance in plants. Oligonucleotides 17:251–257
Ratcliffe FG, MacFarlane SA, Baulcombe DC (1999) Gene silencing without DNA: RNA-mediated cross-protection between viruses. Plant Cell 11:1207–1215
Roberts CS, Rajagopal S, Smith LA, et al (1998) A comprehensive set of modular vectors for advanced manipulations and efficient transformation of plants by both Agrobacterium and direct DNA uptake methods. CAMBIA Vector Release Manual Version 3.5
Sanford JC, Johnston SA (1985) The concept of parasite-derived resistance—deriving resistance genes from the parasite’s own genome. J Theor Biol 113:395–405
Santa Cruz FC, Hull R, Azzam O (2003) Changes in level of virus accumulation and incidence of infection are critical in the characterization of Rice tungro bacilliform virus (RTBV) resistance in rice. Arch Virol 148:1465–1483
Sivamani E, Huet H, Shen P, Ong CA, deKochko A, Fauquet CM, Beachy RN (1999) Rice plants (Oryza sativa L.) containing Rice tungro spherical virus (RTSV) coat protein transgenes are resistant to virus infection. Mol Breed 5:177-185
Tenllado F, Martinez-Garcia B, Vargas M, Dias-Ruiz JR (2003) Crude extracts of bacterially-expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol 3:3
Tougou M, Furutani N, Yamagishi N, Shizukawa Y, Takahata Y, Hidaka S (2006) Development of resistant transgenic soybeans with inverted repeat-coat protein genes of soybean dwarf virus. Plant Cell Rep 25:1213–1218
Vance V, Vaucheret H (2001) RNA silencing in plants—defense and counter-defense. Proc Natl Acad Sci USA 292:2277–2280
Vanitharani R, Chellappan P, Fauquet CM (2003) Short interfering RNA-mediated interference of gene expression and viral DNA accumulation in cultured plant cells. Proc Natl Acad Sci USA 100:9632–9636
Verma V, Dasgupta I (2007) Sequence analysis of the complete genomes of two Rice tungro spherical virus isolates from India. Arch Virol 152:645–648
Wang MB, Upadhyaya NM, Brettel RIS, Waterhouse PM (1997) Intron-mediated improvement of a selectable marker gene for plant transformation using Agrobacterium tumefaciens. J Genet Breed 51:325–334
Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95:13959–13964
Yu H, Kumar PP (2003) Post-transcriptional gene silencing in plants by RNA. Plant Cell Rep 22:167–174
Acknowledgement
Authors acknowledge the expert technical assistance of Shri. Anand Singh Rana and efficient maintenance of plants and insects by Shri. Trilok Singh Rawat This work was generously supported by grants from Department of Biotechnology, Government of India, New Delhi to I. D. and M. V. R. (Grant no. BT/AB/NP/07/98). H. T is grateful to University Grants Commission, New Delhi for the award of senior research fellowship.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tyagi, H., Rajasubramaniam, S., Rajam, M.V. et al. RNA-interference in rice against Rice tungro bacilliform virus results in its decreased accumulation in inoculated rice plants. Transgenic Res 17, 897–904 (2008). https://doi.org/10.1007/s11248-008-9174-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-008-9174-7