Abstract
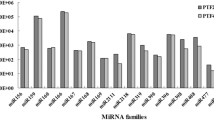
Paulownia fortunei (family Paulowniaceae) is an economically important tree species indigenous to China. Autotetraploid cultivars of P. fortunei have better growth and wood quality than their diploid counterparts. MicroRNAs (miRNAs) play vital regulatory roles in plant growth, development, and biotic and abiotic stress responses by direct cleavage of transcripts, translational repression, or chromatin modification. Although miRNAs have been identified in various plant species, no reports of miRNAs in P. fortunei have been published so far. To study the functions of miRNAs in the autotetraploid P. fortunei, four sequencing libraries from the autotetraploid and its corresponding diploid plants were constructed. 142 conserved miRNAs grouped into 41 families, and 38 novel miRNAs were obtained. Among these miRNAs, 58 were up-regulated and 30 were down-regulated in the autotetraploid relative to the diploid. MiRNA target genes were identified using a degradome sequencing approach and the differently expressed miRNAs and their target genes were validated by real time PCR analysis. To our knowledge, this is the first study to identify conserved and novel miRNAs and their potential targets from diploid and autotetraploid Paulownia plants using high-throughput sequencing and degradome analysis. Our results provide valuable information on P. fortunei miRNAs and their targets, and will help build a foundation for future studies of the biological functions of miRNA-mediated gene regulation in P. fortunei.




Similar content being viewed by others
References
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Struct Bio 8(2):135–141
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18(10):758–762. doi:10.1016/j.cub.2008.04.042
An FM, Chan MT (2012) Transcriptome-wide characterization of miRNA-directed and non-miRNA-directed endonucleolytic cleavage using Degradome analysis under low ambient temperature in Phalaenopsis aphrodite subsp. formosana. Plant Cell Physiol 53(10):1737–1750. doi:10.1093/pcp/pcs118
Anssour S, Krugel T, Sharbel TF, Saluz HP, Bonaventure G, Baldwin IT (2009) Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of Nicotiana attenuata and Nicotiana obtusifolia. Ann Bot 103(8):1207–1217. doi:10.1093/aob/mcp058
Ao Y, Wang Y, Chen L, Wang T, Yu H, Zhang Z (2012) Identification and comparative profiling of microRNAs in wild-type Xanthoceras sorbifolia and its double flower mutant. Genes Genom 34(5):561–568
Axtell MJ, Snyder JA, Bartel DP (2007) Common functions for diverse small RNAs of land plants. Plant cell 19(6):1750–1769. doi:10.1105/tpc.107.051706
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. doi:10.1007/s00425-010-1231-9
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33(20):e179. doi:10.1093/nar/gni178
Chuck G, Candela H, Hake S (2009) Big impacts by small RNAs in plant development. Curr Opin Plant Biol 12(1):81–86
Crawford BC, Nath U, Carpenter R, Coen ES (2004) CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 135(1):244–253
Curley A (1993) Paulownia growing rapidly outside Asia. J Forest 91(6):41
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143(4):1739–1751
Deng M, Dong Y, Zhao Z, Zhang X, Fan G (2013) Llumina-based de novo sequencing and characterization of the transcriptome of Paulownia plant. Sci Silv Sin 49(6):30–36
Fan G, Cao Y, Zhao Z, Yang Z (2007) Induction of autotetraploid of Paulownia fortunei. Sci Silv Sin 43(4):31–35
Fan G, Zhai X, Niu S, Ren Y (2014) Dynamic expression of novel and conserved microRNAs and their targets in diploid and tetraploid of Paulownia tomentosa. Biochimie. doi:10.1016/j.biochi.2014.02.008
Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS (2005) Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol 15(3):331–341
German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, Lu C, Schroth G, Meyers BC, Green PJ (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotech 26(8):941–946. doi:10.1038/nbt1417
Guzman F, Almerao MP, Korbes AP, Loss-Morais G, Margis R (2012) Identification of microRNAs from Eugenia uniflora by high-throughput sequencing and bioinformatics analysis. PLoS ONE 7(11):49811. doi:10.1371/journal.pone.0049811
Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ (2009) Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. PNAS 106(42):17835–17840. doi:10.1073/pnas.0907003106
Hao D, Yang L, Xiao P, Liu M (2012) Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis. Physiol Plant 146(4):388–403. doi:10.1111/j.1399-3054.2012.01668.x
Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P (1994) Fast folding and comparison of RNA secondary structures. Monatsh Chem 125(2):167–188
Khan Y, Yadav A, Bonthala V, Muthamilarasan M, Yadav C, Prasad M (2014) Comprehensive genome-wide identification and expression profiling of foxtail millet [Setaria italica (L.)] miRNAs in response to abiotic stress and development of miRNA database. Plant Cell Tiss Organ Cult 1–14. doi:10.1007/s11240-014-0480-x
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Leitch IJ, Bennett MD (1997) Polyploidy in angiosperms. Trends Plant Sci 2(12):470–476
Li B, Yin W, Xia X (2009) Identification of microRNAs and their targets from Populus euphratica. Biochem Biophys Res Commun 388(2):272–277. doi:10.1016/j.bbrc.2009.07.161
Li Y, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell M, Zhang W, Sunkar R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62(5):742–759. doi:10.1111/j.1365-313X.2010.04187.x
Li B, Qin Y, Duan H, Yin W, Xia X (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot 62(11):3765–3779
Li X, Yu E, Fan C, Zhang C, Fu T, Zhou Y (2012) Developmental, cytological and transcriptional analysis of autotetraploid Arabidopsis. Planta 236(2):579–596. doi:10.1007/s00425-012-1629-7
Li W, Zhang S, Han S, Wu T, Zhang J, Qi L (2013) Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.). Plant Cell Tiss Organ Cult 113(1):131–136
Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408
Lu S, Sun Y, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress–responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17(8):2186–2203
Lu S, Sun Y, Amerson H, Chiang V (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J 51(6):1077–1098. doi:10.1111/j.1365-313X.2007.03208.x
Mackowiak SD (2011) Identification of novel and known miRNAs in deep‐sequencing data with miRDeep2. Curr Protoc Bioinform 36:121011–121015
Meyers B, Axtell M, Bartel B, Bartel D, Baulcombe D, Bowman J, Cao X, Carrington J, Chen X, Green P (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20(12):3186–3190
Miyashima S, Koi S, Hashimoto T, Nakajima K (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138(11):2303–2313
Morin R, O’Connor M, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA (2008) Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18(4):610–621. doi:10.1101/gr.7179508
Moxon S, Jing R, Szittya G, Schwach F, Pilcher RLR, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18(10):1602–1609
Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, Ecker JR, Reed JW (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132(18):4107–4118. doi:10.1242/dev.01955
Nath U, Crawford BC, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299(5611):1404–1407
Niu S, Fan G, Zhao Z, Deng M, Dong Y (2013) MicroRNAs of Paulownia plants and their functional prediction. Sci Silv Sin 49(11):77–82
Paul S, Kundu A, Pal A (2011) Identification and validation of conserved microRNAs along with their differential expression in roots of Vigna unguiculata grown under salt stress[J]. Plant Cell Tiss Organ Cult 105(2):233–242
Ren Y, Chen L, Zhang Y, Kang X, Zhang Z, Wang Y (2012) Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct Integr Genomics 12(2):327–339. doi:10.1007/s10142-012-0271-6
Rhoades M, Reinhart B, Lim L, Burge C, Bartel B, Bartel D (2002) Prediction of plant microRNA targets. Cell 110(4):513–520
Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6(9):230. doi:10.1371/journal.pbio.0060230
Shen H, He H, Li J, Chen W, Wang X, Guo L, Peng Z, He G, Zhong S, Qi Y, Terzaghi W, Deng XW (2012) Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24(3):875–892. doi:10.1105/tpc.111.094870
Shleizer-Burko S, Burko Y, Ben-Herzel O, Ori N (2011) Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138(4):695–704. doi:10.1242/dev.056770
Shu C (1989) A survey of the chromosome number of four species of Paulownia. J Henan Agric Univ 19(1):48–50
Simon SA, Meyers BC (2011) Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol 14(2):148–155
Song C, Wang C, Zhang C, Korir NK, Yu H, Ma Z, Fang J (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genom 11:431. doi:10.1186/1471-2164-11-431
Song Q, Liu Y, Hu X, Zhang W, Ma B, Chen S, Zhang J (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol 11:5. doi:10.1186/1471-2229-11-5
Sun C, Zhao Q, Liu D, You C, Hao Y (2013) Ectopic expression of the apple Md-miRNA156 h gene regulates flower and fruit development in Arabidopsis. Plant Cell Tiss Organ Cult 112(3):343–351
Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu J-K (2008) Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8(1):25
Unver T, Budak H (2009) Conserved microRNAs and their targets in model grass species Brachypodium distachyon. Planta 230(4):659–669
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136(4):669–687
Wan L, Wang F, Guo X, Lu S, Qiu Z, Zhao Y, Zhang H, Lin J (2012) Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol 12:146. doi:10.1186/1471-2229-12-146
Wang Z, Huang J, Huang Y, Li Z, Zheng B (2012) Discovery and profiling of novel and conserved microRNAs during flower development in Carya cathayensis via deep sequencing. Planta 236(2):613–621. doi:10.1007/s00425-012-1634-x
Wei M, Wei H, Wu M, Song M, Zhang J, Yu J, Fan S, Yu S (2013) Comparative expression profiling of miRNA during anther development in genetic male sterile and wild type cotton. BMC Plant Biol 13:66. doi:10.1186/1471-2229-13-66
Xie F, Frazier TP, Zhang B (2011) Identification, characterization and expression analysis of MicroRNAs and their targets in the potato (Solanum tuberosum). Gene 473(1):8–22
Yakovlev IA, Fossdal CG, Johnsen O (2010) MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol 187(4):1154–1169. doi:10.1111/j.1469-8137.2010.03341.x
Zhai X, Zhang X, Zhao Z, Deng M, Fan G (2012) Study on wood physical properties of tetraploid Paulownia fortunei. J Henan Agric Univ 46(6):2012
Zhang B, Pan X, Cobb GP, Anderson TA (2006a) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289(1):3–16
Zhang J, Guo W, Deng X (2006b) Relationship between ploidy variation of citrus calli and competence for somatic embryogenesis. Acta Genet Sin 33(7):647–654
Zhang B, Pan X, Anderson T (2006c) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhang X, Zhai X, Fan G, Deng M, Zhao Z (2012) Observation on microstructure of leaves and stress tolerance analysis of different tetraploid Paulownia. J Henan Agric Univ 46(6):646–650
Zhang X, Fan G, Zhao Z, Cao X, Zhao G, Deng M, Dong Y (2013) Analysis of diploid and its autotetraploid Paulownia tomentosa × Paulownia fortunei with AFLP and MSAP. Sci Silv Sin 10:026
Zhang J, Zhang S, Li S, Han S, Li W, Li X, Qi L (2014) Regulation of synchronism by abscisic-acid-responsive small noncoding RNAs during somatic embryogenesis in larch (Larix leptolepis). Plant Cell Tiss Organ Cult 116(3):361–370
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Grant No. 30271082, 30571496, U1204309), by the Outstanding Talents Project of Henan Province (Grant No. 122101110700), by the Transformation Project of the National Agricultural Scientific and Technological Achievement of China (Grant No. 2012GB2D000271), and by Science and Technology Innovation Team Project of Zhengzhou City (Grant No. 121PCXTD515).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11240_2014_546_MOESM4_ESM.doc
Online Resource 4 The secondary structures of novel P. fortunei miRNA precursors. (The mature miRNAs are in red, and the miRNA*s are in green) (DOC 69 kb)
11240_2014_546_MOESM6_ESM.doc
Online Resource 6 Species distribution of the BLAST matches of the degradome targets. This figure shows the distributions of unigenes BLASTX matches against the nr protein database (cutoff value E < 10−5) and the proportions for each species (DOC 116 kb)
11240_2014_546_MOESM7_ESM.doc
Online Resource 7 GO analyses of the targets of the conserved and novel miRNAs in P. fortunei. Blue bar, indicate the Biological process; Red bar, indicate the Cellular component; Green bar, indicate the Molecular function (DOC 140 kb)
Rights and permissions
About this article
Cite this article
Niu, S., Fan, G., Zhao, Z. et al. High-throughput sequencing and degradome analysis reveal microRNA differential expression profiles and their targets in Paulownia fortunei . Plant Cell Tiss Organ Cult 119, 457–468 (2014). https://doi.org/10.1007/s11240-014-0546-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-014-0546-9