Abstract
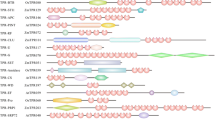
The Arabidopsis thaliana PRP39, encoding a tetratricopeptide repeat protein, is a novel gene that affects flowering time via an autonomous pathway. In this study, a PRP39 homolog gene was isolated from Doritaenopsis ‘Tinny Tender’ (Doritaenopsis Happy smile × Happy valentine) and designated as DhPRP39. The full-length DhPRP39 cDNA was 2,962 bp long with a 2,472 bp open reading frame and encodes 823 amino acids. The putative DhPRP39-encoded protein contained four conserved, three half-a-tetratricopeptide repeat, and one RNA14 regions. Amino acid sequence alignment showed that DhPRP39 shares a high similarity to PRP39 homologs from other species. Real-time quantitative reverse transcription polymerase chain reaction analysis showed that DhPRP39 is ubiquitously expressed in vegetative and reproductive organs. The transcripts of this gene reached higher levels in the vegetative organs (roots, stems, and leaves) during the transition from vegetative to reproductive growth, in which the stems exhibited the strongest expression. DhPRP39 was overexpressed in Arabidopsis thaliana, and flowering of these transgenic plants were much delayed by 6–7 days compared with wild-type Arabidopsis. DhPRP39 may play an important role in the regulation of plant flowering.






Similar content being viewed by others
Abbreviations
- CaMV:
-
Cauliflower mosaic virus
- gDNA:
-
Genomic DNA
- HAT:
-
Half-a-TPR
- RACE:
-
Rapid amplification of cDNA ends
- ORF:
-
Open reading frame
- PPR:
-
Pentatricopeptide repeat
- SSH:
-
Suppression subtractive hybridization
- TPR:
-
Tetratricopeptide repeat
- WT:
-
Wild-type
References
Bezerra IC, Michaels SD, Schomburg FM, Amasino RM (2004) Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J 40:112–119
Blatch GL, Lässle M (1999) The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. Bio Essays 21:932–939
Blom E, van de Vrugt HJ, de Vries Y, de Winter JP, Arwert F, Joenje H (2004) Multiple TPR motifs characterize the Fanconi anemia FANCG protein. DNA Repair 3:77–84
Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16:S18–S31
Boudreau E, Nickelsen J, Lemaire SD, Ossenbuhl F, Rochaix JD (2000) The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J 19:3366–3376
Chen WH, Tseng YC, Liu YC, Chuo CM, Chen PT, Tseng KM, Yeh YC, Ger MJ, Wang HL (2008) Cool-night temperature induces spike emergence and affects photosynthetic efficiency and metabolizable carbohydrate and organic acid pools in Phalaenopsis aphrodite. Plant Cell Rep 27:1667–1675
Chung S, Zhou Z, Huddleston KA, Harrison DA, Harrison DA, Reed R, Colemanc TA, Rymond BC (2002) Crooked neck is a component of the human spliceosome and implicated in the splicing process. Biochim Biophys Acta 1576:287–297
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Cui YY, Pandey DM, Hahn EJ, Park KY (2004) Effect of drought on physiological aspects of crassulacean acid metabolism in Doritaenopsis. Plant Sci 167:1219–1226
D’Andrea LD, Regan L (2003) TPR proteins: the versatile helix. Trends Biochem Sci 28:655–662
Das AK, Cohen PW, Barford D (1998) The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein–protein interactions. EMBO J 17:1192–1199
Guyomarc’h S, Vernoux T, Traas J, Zhou DX, Delarue M (2004) MGOUN3, an Arabidopsis gene with TetratricoPeptide-Repeat-related motifs, regulates meristem cellular organization. J Exp Bot 55:673–684
He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302:1751–1754
Jack T (2004) Molecular and genetic mechanisms of floral control. Plant Cell 16:S1–S17
Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290:344–347
Kiselev KV, Turlenko AV, Zhuravlev YN (2010) Structure and expression profiling of a novel calcium-dependent protein kinase gene PgCDPK1a in roots, leaves, and cell cultures of Panax ginseng. PCTOC 103:197–204
Koornneef M, Alonso-Blanco C, Peeters AJ, Soppe W (1998) Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol 49:345–370
Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433:326–330
Lee HC, Chen YJ, Markhart AH, Lin TY (2007) Temperature effects on systemic endoreduplication in orchid during floral development. Plant Sci 172:588–595
Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Kim J, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16:731–740
Lockhart SR, Rymond BC (1994) Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol Cell Biol 14:3623–3633
Luo X, Zhang C, Sun X, Qin Q, Zhou M, Paek KY, Cui Y (2011) Isolation and characterization of a Doritaenopsis hybrid GIGANTEA gene, which possibly involved in inflorescence initiation at low temperatures. Kor J Hort Sci Technol 29(2):135–143
Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89:737–745
Macknight R, Duroux M, Laurie R, Dijkwel P, Simpson G, Dean C (2002) Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14:877–888
McLean MR, Rymond BC (1998) Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol Cell Biol 18:353–360
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13:935–941
Mouradov A, Creme F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14:S111–S130
Preker PJ, Keller W (1998) The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem Sci 23:15–16
Qin Q, Kaas Q, Zhang C, Zhou L, Luo X, Zhou M, Sun X, Zhang L, Paek KY, Cui Y (2011) The cold awakening of Doritaenopsis ‘Tinny Tender’ orchid flowers: the role of leaves in cold-induced bud dormancy release. J Plant Growth Regul. doi:10.1007/s00344-011-9226-8
Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13:1427–1436
Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). P Natl Acad Sci USA 97(6):3753–3758
Shen L, Chen Y, Su X, Zhang S, Pan H, Huang M (2012) Two FT orthologs from Populus simonii Carrière induce early flowering in Arabidopsis and poplar trees. PCTOC 108:371–379
Simpson GG, Dean C (2002) Arabidopsis, the rosetta stone of flowering time? Science 296:285–289
Simpson GG, Gendall AR, Dean C (1999) When to switch to flowering. Annu Rev Cell Dev Biol 15:519–550
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113:777–787
Small ID, Peeters N (2000) The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25:46–47
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Wang C, Tian Q, Hou Z, Mucha M, Aukerman M, Olsen OA (2007) The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep 26:1357–1366
Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ (2010) Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J 8:749–771
Xuan N, Jin Y, Zhang H, Xie Y, Liu Y, Wang G (2011) A putative maize zinc-finger protein gene, ZmAN13, participates in abiotic stress response. PCTOC 107:101–112
Zakizadeh H, Stummann BM, Lütken H, Müller R (2010) Isolation and characterization of four somatic embryogenesis receptor-like kinase (RhSERK) genes from miniature potted rose (Rosa hybrida cv. Linda). PCTOC 101:331–338
Zhu Y, Zhao HF, Ren GD, Yu XF, Cao SQ, Kuai BK (2005) Characterization of a novel developmentally retarded mutant (drm1) associated with the autonomous flowering pathway in Arabidopsis. Cell Res 15(2):133–140
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Nos. 30771762 and 31170658) and the Zhejiang Provincial Natural Science Foundation of China (Grant No. Y3090532).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, X., Qin, Q., Zhang, J. et al. Cloning and characterization of a Doritaenopsis hybrid PRP39 gene involved in flowering time. Plant Cell Tiss Organ Cult 110, 347–357 (2012). https://doi.org/10.1007/s11240-012-0156-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-012-0156-3