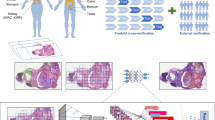
Abstract
Diagnosis of lymph node metastases is a challenging task for pathologists, involving an extensive screening of the pathological scans. Automating diagnostic processes reduces the workload of pathologists and yields high accuracy by the virtue of advances in technology. In this study, a novel ensemble-based framework is proposed for the classification of lymph node metastases. The proposed ensemble framework comprises different pre-trained CNN models such as DenseNet201, InceptionV3 and ResNeXt-50. In the proposed framework, an attention fusion network is utilized to amalgamate the predictions of the individual models. The proposed framework achieves an AUC-ROC of 0.9816 which surpasses the highest AUC-ROC achieved by the conventional approaches on the PCam benchmark dataset.









Similar content being viewed by others
References
Chalbatani GM, Dana H, Memari F, Gharagozlou E, Ashjaei S, Kheirandish P, Marmari V, Mahmoudzadeh H, Mozayani F, Maleki AR et al (2019) Biological function and molecular mechanism of piRNA in cancer. Practical Lab Med 13:e00113
Bejnordi BE, Veta M, Van Diest PJ, Van Ginneken B, Karssemeijer N, Litjens G, Van Der Laak JA, Hermsen M, Manson QF, Balkenhol M et al (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22):2199–2210
Nci-dictionaries. URL https://www.cancer.gov/publications/dictionaries/cancer-terms/def/metastasis. Accessed 22 Mar 2020
Li C, Wang X, Liu W, Latecki LJ, Wang B, Huang J (2019) Weakly supervised mitosis detection in breast histopathology images using concentric loss. Med Image Anal 53:165–178
Kassani SH, Kassani PH, Wesolowski MJ, Schneider KA, Deters R (2019) Classification of histopathological biopsy images using ensemble of deep learning networks. arXiv preprint arXiv:1909.11870
Litjens G, Kooi T, Bejnordi BE, Setio AA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Liu Y, Gadepalli K, Norouzi M, Dahl GE, Kohlberger T, Boyko A, Venugopalan S, Timofeev A, Nelson PQ, Corrado GS, et al. (2017) Detecting cancer metastases on gigapixel pathology images. arXiv preprint arXiv:1703.02442
Veeling BS, Linmans J, Winkens J, Cohen T, Welling M (2018) Rotation equivariant CNNS for digital pathology. In: International Conference on Medical Image Computing and Computer-assisted Intervention, pp 210–218. Springer
Pang H, Lin W, Wang C, Zhao C (2018) Using transfer learning to detect breast cancer without network training. In: 2018 5th IEEE International Conference on Cloud Computing and Intelligence Systems (CCIS), pp 381–385. IEEE
Jaiswal AK, Panshin I, Shulkin D, Aneja N, Abramov S (2019) Semi-supervised learning for cancer detection of lymph node metastases. arXiv preprint arXiv:1906.09587
Teh EW, Taylor GW (2019) Learning with less data via weakly labeled patch classification in digital pathology. arXiv, pages arXiv–1911
Palatnik de Sousa I, Maria Bernardes Rebuzzi Vellasco M, Costa da Silva E (2019) Local interpretable model-agnostic explanations for classification of lymph node metastases. Sensors 19(13):2969
Burcak KC, Baykan ÖK, Uğuz H (2020) A new deep convolutional neural network model for classifying breast cancer histopathological images and the hyperparameter optimisation of the proposed model. J Supercomput. https://doi.org/10.1007/s11227-020-03321-y
Acharya S, Alsadoon A, Prasad PW, Abdullah S, Deva A (2020) Deep convolutional network for breast cancer classification: enhanced loss function (ELF). J Supercomput. https://doi.org/10.1007/s11227-020-03157-6
Khamparia A, Gupta D, de Albuquerque VH, Sangaiah AK, Jhaveri RH (2020) Internet of health things-driven deep learning system for detection and classification of cervical cells using transfer learning. J Supercomput. https://doi.org/10.1007/s11227-020-03159-4
Zhou L-Q, Xing-Long W, Huang S-Y, Ge-Ge W, Ye H-R, Wei Q, Bao L-Y et al (2020) Lymph node metastasis prediction from primary breast cancer us images using deep learning. Radiology 294(1):19–28
Sun Q, Lin X, Zhao Y, Li L, Yan K, Liang D, Sun D, Li ZC (2020) Deep learning vs. radiomics for predicting axillary lymph node metastasis of breast cancer using ultrasound images: don’t forget the peritumoral region. Front Oncol 10:53
Sadeghi M, Maldonado I, Abele N, Haybaeck J, Boese A, Poudel P, Friebe M (2019) Feedback-based self-improving CNN algorithm for breast cancer lymph node metastasis detection in real clinical environment. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 7212–7215. IEEE
Kumar M, Alshehri M, AlGhamdi R, Sharma P, Deep V (2020) A DE-ANN inspired skin cancer detection approach using fuzzy c-means clustering. Mobile Netw Appl 25:1319–1329
Goyal V, Singh G, Tiwari OM, Punia SK, Kumar M (2019) Intelligent skin cancer detection mobile application using convolution neural network. J Adv Res Dyn Control Syst 11(7(SI)):253–259
Vo DM, Nguyen NQ, Lee SW (2019) Classification of breast cancer histology images using incremental boosting convolution networks. Inf Sci 482:123–138
Bejnordi BE, Zuidhof G, Balkenhol M, Hermsen M, Bult P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak J (2017) Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images. J Med Imag 4(4):044504
Nahid AA, Mehrabi MA, Kong Y (2018) Histopathological breast cancer image classification by deep neural network techniques guided by local clustering. BioMed Res Int. https://doi.org/10.1155/2018/2362108
Alom MZ, Yakopcic C, Nasrin MS, Taha TM, Asari VK (2019) Breast cancer classification from histopathological images with inception recurrent residual convolutional neural network. J Digital Imaging 32(4):605–617
Bas Veeling. Pcam-dataset. URL https://www.kaggle.com/c/histopathologic-cancer-detection. Accessed 15 Jan 2020
Qureshi SA, Saha S, Hasanuzzaman M, Dias G (2019) Multitask representation learning for multimodal estimation of depression level. IEEE Intell Syst 34(5):45–52
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826
Xie S, Girshick R, Dollár P, Tu Z, He K (2017) Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1492–1500
Smith LN (2018) A disciplined approach to neural network hyper-parameters: part 1–learning rate, batch size, momentum, and weight decay. arXiv preprint arXiv:1803.09820
Ho Y, Wookey S (2019) The real-world-weight cross-entropy loss function: modeling the costs of mislabeling. IEEE Access 8:4806–4813
Balles L, Romero J, Hennig P (2016) Coupling adaptive batch sizes with learning rates. arXiv preprint arXiv:1612.05086
Poria S, Cambria E, Hazarika D, Mazumder N, Zadeh A, Morency LP (2017) Multi-level multiple attentions for contextual multimodal sentiment analysis. In: 2017 IEEE International Conference on Data Mining (ICDM), pp. 1033–1038. IEEE
Wang G, Li W, Ourselin S, Vercauteren T (2018) Automatic brain tumor segmentation using convolutional neural networks with test-time augmentation. In: International MICCAI Brainlesion Workshop, pp. 61–72. Springer
Matsunaga K, Hamada A, Minagawa A, Koga H (2017) Image classification of melanoma, nevus and seborrheic keratosis by deep neural network ensemble. arXiv preprint arXiv:1703.03108
Jin H, Li Z, Tong R, Lin L (2018) A deep 3D residual CNN for false-positive reduction in pulmonary nodule detection. Med Phys 45(5):2097–2107
Nalepa J, Myller M, Kawulok M (2019) Training-and test-time data augmentation for hyperspectral image segmentation. IEEE Geosci Remote Sens Lett 17(2):292–296
Wang G, Li W, Aertsen M, Deprest J, Ourselin S, Vercauteren T (2019) Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing 338:34–45
Waibel DJ, Boushehri SS, Marr C (2020) Instantdl-an easy-to-use deep learning pipeline for image segmentation and classification. bioRxiv
Kandel I, Castelli M (2020) How deeply to fine-tune a convolutional neural network: a case study using a histopathology dataset. Appl Sci 10(10):3359
Duan Y, Sun L, Wang Y (2019) Se-densenet: attention-based network for detecting pathological images of metastatic breast cancer. In: Proceedings of the 2019 8th International Conference on Computing and Pattern Recognition, pp. 240–245
Liu M, Yu Y, Liao Q, Zhang J (2020) Histopathologic cancer detection by dense-attention network with incorporation of prior knowledge. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pp. 466–470. IEEE
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rane, C., Mehrotra, R., Bhattacharyya, S. et al. A novel attention fusion network-based framework to ensemble the predictions of CNNs for lymph node metastasis detection. J Supercomput 77, 4201–4220 (2021). https://doi.org/10.1007/s11227-020-03432-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-020-03432-6