Abstract
Association constants of 2,6-bis(alkylcarbonylamino)pyridines (alkyl = methyl or ethyl) and their perfluoroalkyl analogues with succin- and maleimide as well as with 2,2′-dipyridylamine (complementary DAD and ADA hydrogen bonding motifs are responsible for formation of the associates) have been determined by NMR titrations and quantum chemical calculations. Interactions of 2,6-bis(alkylcarbonylamino)pyridines with imides differ by character from these of perfluoroalkyl analogues. Such large difference was not observed for the 2,2′-dipyridylamine associates. Since fluorine atoms cause carbonylamino groups to be stronger hydrogen bond donors, perfluorinated species of this type were found to be more stable. Single crystal X-ray structures of 2,6-bis(trifluoromethylcarbonylamino)pyridine and 2,6-bis(pentafluoroethylcarbonylamino)pyridine have been also determined.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Intermolecular multiple hydrogen bonding (HB’ing) is of major importance in biochemistry and supramolecular chemistry. It plays a crucial role in formation of the double helix of DNA [1] and in action of artificial receptors used in biochemistry [2, 3]. Hydrogen bonding is the most common non-covalent interaction [2, 4] observed in biosensing, self-complementary aggregation, and non-covalent polymer formation [5–7]. Significance of hydrogen bonding is based on its directionality and reversibility [6]. There are few factors that influence the HB’ed complex stability. These are number of hydrogen bonds, hydrogen bonding pattern [8, 9], secondary interactions [10, 11]. Also the tautomeric equilibrium may be responsible for forming variously stabilized associates. This is due to group and HB’ing pattern changes upon proton transfer.
It is known that secondary interactions result in strengthening of the AAAA/DDDD-type [12] association in comparison with that of ADAD/DADA [13]. The same is true for AAA/DDD [14–17] versus DDA/AAD [18, 19] versus ADA/DAD pairs [7, 8, 20–22]. Although the ADA and the DAD motifs are not self-complementary, their self-association is still possible due to rotamerism (Scheme 1) or tautomerism [23–26]. On the other hand, 2,6-bis(acrylamido)pyridine does not form dimers [27].
Steric hindrance [28–31] weakens basicity of 2,6-di-tert-butylpyridine [32]. Thus, the parallel effect should be observed for non-covalent interactions. Also the electronic repulsion is believed to be responsible for weakening of the association of bis(acetylamino)triazine derivatives with imides [33].
The organized structures of 2,6-diacetylaminopyridines with imides were obtained by their aggregation on the Ag surface [34]. Hydrogen bonding in these compounds was studied mainly from the point of view of binding in nucleic bases [35–37], bisurea derivatives [38], structures stabilized by π–π interactions [39], receptors for barbituric acid [40], and fluorine anion [41], enhancement of the fluorescence of conjugated polymeric chain upon complexation [42], cooperative catalyst [43], and potentially drug delivery nanomaterials [44]. Hydrogen bonding and possibility of the complex formation in solution and in solid state between the 2,6-diacetylaminopyridine and enolate derivative of barbituric acid has been shown recently by the X-ray diffraction [45]. Interestingly, conformation of the amide group in N-(pyrimidin-2-yl)pentafluorobenzamide derivatives was found to be of the cis type, especially when the 2,6-diacetylaminopyridine derivative stabilized this form by hydrogen bonding [46].
Feibush have suggests that 2,6-bis(pivaloylamino)pyridine does not form complexes with imides due to steric crowding [47]. On the other hand, stronger hydrogen bonding capability of proton in the NHCOCF3 group (as compared to that in NHCOCH3) is expected to strengthen the complex stability. 2,6-Bis(trifluoromethylcarbonylamino)pyridine has been investigated as a hydrogen bonding counterpart stabilizing the flavin radical anion [48] and as a hydrogen bonding receptor for barbiturate [49]. The acylation of amino moiety, however, not always led to the greater stability of non-covalent associates [33]. It was suggested that this is due to electronic repulsion/secondary repulsive interactions.
Imides are complementary by hydrogen bonding to with 2,6-bis(alkylcarbonylamino)pyridines. The triple hydrogen bonds are expected to strengthen the association especially with the perfluoroalkyl derivatives. Clarification of these interactions between succin- and maleimide, 2,2′-dipyridynamine and 2,6-bis(alkylcarbonylamino)pyridines is the main goal of this article.
Results and discussion
Formulas of 2,6-bis(acylamino)pyridines 1–4 with 2,2′-dipyridylamine (5) and imides (6) and (7) as well as numbering of positions in their molecules are depicted in Fig. 1.
NMR
Since NMR chemical shifts of the NH/OH protons are sensitive to the concentration and solvent properties, the 1H NMR spectra of all neat compounds and their 1:1 mixtures were recorded at the same concentration (see “Experimental”). On the other hand, their 13C and 15N spectra were run for the saturated solutions. The chemical shifts are available in the Supporting Information Section (SI, Table S1). H7 protons in 2 and 4 were found to be more acidic than these in 1 and 3. Perfluorination of the alkyl groups results in deshielding of the 1H7 signal by 0.7–0.8 ppm.
The complexation induced shift (CIS) values of the amide protons (δ(H7)) for 1–4 and their 1:1 mixtures with 2,2′-dipyridylamine (5), succinimide (6), and maleimide (7) are collected in Table 1 (see also Table S1).
The association constants (Table 1) show that the most stable complexes are 1 + 6 and 3 + 6. It is noteworthy that K assoc obtained now are comparable to these for other triple hydrogen-bonded systems [44, 50–52].
Based on the CIS and K assoc values for mixtures of 1–4 with 5 (Table 1) and δ(H7) for the neat 1–4 (Table S1) one can see that perfluoroalkyl groups increase the hydrogen bond donor properties of H7. This results in increasing stabilities of complexes carrying the electron acceptor groups. Contrary to 1 and 3, negligible effect of complexation of succin- and maleimide was found for perfluoro analogues 2 and 4 implying that their association with those imides is very weak. Such weak complexation was earlier observed in mixture of 2 with flavin [48]. An explanation for the weak association of 2 and 4 with 6 and 7 can be that CF3 and C2F5 groups create electronic repulsions towards the hydrogen bond acceptors, i.e., carbonyl oxygens of the imide. Differing from that in 2,2′-dipyridilamine partial rotation of the pyridine ring around the N1–C2 bond [53] (Fig. 2) causes that this molecule can adopt a geometry that allows complex formation even when substituents show some repulsion with the pyridine rings of 5.
Owing to low rotation barrier of the perfluoroalkyl groups around the C–N bond in the amide [46], both the trans and cis forms (Scheme 2) may be expected to be present in the complexes of 2 and 4 with imides 6 and 7. One should keep in mind, however, that electronic repulsion between the carbonyl oxygen of 6 and 7 and fluorine or oxygen atoms of 2 and 4 may destabilize the said complexes (Scheme 3).
Additional 1H NMR experiments were also run with three component mixtures to clarify what happens in case of competitive binding. The detailed results and discussion can be found in SI.
X-ray structural data
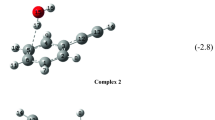
Single crystal structures of 2 and 4 (Fig. 3) show that all the substituent C atoms (except these of CF3 groups in 4) lie almost in the plane of pyridine ring. Although two-dimensional schematic drawings of 2 and 4 indicate these compounds to be symmetrical, there is no crystallographic mirror plane or other symmetry element found along the N1···C4–H4 axis. Despite the amide side chains in 2,6-bis(acylamino)pyridines are slightly twisted with respect to the pyridine ring (Table S12) their geometries are comparable. Figure 2 shows molecular structures of 2 and 4 as the ORTEP-diagrams [54]. Compounds 2 and 4 are held in the crystal phase by a net of the intermolecular hydrogen bonds (Fig. S1, for distances N7···O9 and N(R2)···O(R2) see Table S12).
The ORTEP-diagrams [54] of the compounds 2 (upper) and 4 with thermal ellipsoids drawn at 50% probability level (heteroatoms displayed as octant shaded model)
Calculations
Molecular geometries were calculated using the DFT method (M05). It is less time-invasive than, for example, calculations at the MP2 level. The M05 functional is optimized for calculation of many types of non-covalent interactions. This methodology has been previously compared [55] by us with B3LYP and MP2 ones for non-covalent intermolecular interactions in 1,8-naphthyridine derivatives. Detailed geometry data for complexes are collected in SI (Tables S3–S9).
Geometries of the optimized complexes support the results obtained by 1H NMR spectrometry. Substitution of H by F atoms in the acylamino moieties makes H7 protons more acidic. The H7···X3′ distance (X3′ is the nitrogen and oxygen atom in 5 and 6 (and 7), respectively) shows that hydrogen bond is always shorter in the fluorinated derivatives. However, there are some exceptions: H7···X3′ distances in 4 + 6 and in 3 + 6 are practically identical. Moreover, H7 in 4 + 7 is more faraway from X3′ than in 3 + 7. As a result of shortening of the H7···X3′ distance, the H1′···N1 hydrogen bonds in 2 + 5 and 4 + 5 complexes are noticeably shorter than these in 1 + 5 and 3 + 5. The X3′···C8 and X3′···C(R3) distances are noticeably larger in the complexes carrying the CF3 and C2F5 groups. Lower values of C2–N7–C8 angle (Table S7) in fluoroalkyl as compared to these in alkyl derivatives suggest that repulsion between the pyridine ring of 5 or oxygen atoms of imides and fluorine atoms of 2 and 4 takes place. It is noteworthy that energy of the complex formation for fluorinated derivatives 2 and 4 with imides 6 and 7 is ca. 18.4 ± 1 kJ/mol lower than that of 1 and 3 with the same imides (Table S10).
The intermolecular interactions influence electron distribution in each complex. The orbital contours of 1 + 6 and 2 + 6 show the interaction of the hydrogen bond character (exemplified on Fig. 4). Other orbitals (HOMO-20 and HOMO-14, HOMO-27 (no H-bond is visible at this contour level), and HOMO-24, HOMO-29 and HOMO-30, HOMO-35 and HOMO-41, HOMO-36 and HOMO-42) involved in hydrogen bonding in 1 + 6 and 2 + 6 were also considered. There are two orbitals in 2 + 6 (HOMO-38 and HOMO-40) that show the electron repulsion is present. No such orbitals were found for the 1 + 6 complex. Orbital contours are collected in the SI.
Conclusions
Complexes of 2,6-bis(alkylcarbonylamino)pyridines with succin- and maleimide as well as with 2,2′-dipyridylamine are stabilized by the triple hydrogen bonds. Substitution of H by F in the alkyl parts of 2,6-bis(alkylcarbonylamino)-pyridines makes the amide protons better hydrogen bond donor. On the other hand, strong intermolecular CO/F electronic repulsion diminishes efficiency of these compounds to associate with imides whereas the conformational flexibility of 2,2′-dipyridilamine enables its association with 2,6-diacylaminopyridines. The association constants of imides and 2,2′-dipyridilamine with 2,6-diacylaminopyridines follow the concept of steric repulsion. The spectra of the double (1 + 4 and 1 + 6) versus triple mixtures (1 + 4 + 6) studied by 1H NMR confirm complexes of imides with 2,6-bis(alkylcarbonylamino)pyridines to be much more stable than these with 2,6-bis(perfluoroalkylcarbonylamino)pyridines, showing the selective binding of imides by non-fluorinated derivatives. The linear dependence between the association constants and the complexation induced shifts enables the latter to be used as a preliminary probe for relative complex stability. The agreement of the computational data (geometry, energy and visualization of molecular orbitals) with experimental one, including the electronic repulsion between oxygen and fluorine, suggests that DFT method is able to describe hydrogen bonding and electronic repulsion reliably.
Experimental
Synthesis
Compounds 1–4 were obtained by refluxing (2 h) of the mixture of 2,6-diaminopyridine (0.2 g, 1.8 mmol) and 2 mL of the appropriate acid anhydride. Excess of the latter compound was decomposed by addition of water (10 mL) and saturated aqueous sodium carbonate solution (5 mL). The obtained mixture was then extracted with chloroform (2 × 15 mL), organic layer dried (Na2SO4), and evaporated to dryness under reduced pressure. The crude products were further purified by recrystallization. Melting points: 1, 199–202 °C (C6H14/AcOEt, white powder) (lit. 202–203 °C [56], 205–206 °C [57]), 2, 154–158 °C (C6H14/AcOEt, pale-yellow needles), 3, 127–128 °C (C6H14/AcOEt, white powder) and 4, 105–107 °C (C6H14/AcOEt, pale-brown crystals). Satisfactory elemental analytical data were obtained for synthesized compounds, i.e., 1 calcd C9H11N3O2 C 55.95, H 5.74, N 21.75, found: C 55.68, H 5.70, N 21.52, 2 calcd C11H15N3O2 C 59.71, H 6.83, N 18.99 found: C 59.54, H 6.76, N 18.69, 3 calcd C9H5F6N3O2 C 35.90, H 1.67, N 13.95 found: C 35.73, H 1.64, N 13.78, 4 calcd C11H5F10N3O2 C 32.93, H 1.26, N 10.47 found: C 32.65, H 1.22, N 10.27. Compounds 5–7 were commercially available and were used as obtained after drying in desiccator.
NMR
1H NMR experiments were run with a Bruker Avance DRX 500 spectrometer equipped with an inverse detection 5-mm diameter probehead with a z-gradient for equimolar CDCl3 solutions at 303 K. 1H and 13C NMR chemical shifts are referenced to an internal TMS (δ = 0.00 ppm). Owing to the limited solubility, 13C and 15N spectra of all compounds are run for their saturated solutions. Acquisition and processing parameters are the same as reported earlier [58]. The 2D pulsed field z-gradient (PFG) selected 1H,13C HMQC, and 1H,13C HMBC experiments were run to assign reliably the 13C NMR spectra [58]. 15N NMR chemical shifts (referenced to an external neat 15N-natural abundance nitromethane, δ = 0.0 ppm) are those obtained with the PFG 1H, 15N HMBC experiments [58].
Association experiments
Equimolar quantities (0.089 mmol) of 1 (17.2 mg), 2 (26.9 mg), 3 (19.7 mg), 4 (35.7 mg), 5 (15.3 mg), 6 (8.8 mg), and 7 (8.6 mg) were dissolved in acetone (10 mL). Solutions of 1–4 (3 mL) were then combined with solutions of 5–7 (3 mL) to obtain the 1 + 5, 1 + 6, 1 + 7, 2 + 5, 2 + 6, 2 + 7, 3 + 5, 3 + 6, and 3 + 7 complexes. Additional 3 mL solutions of compounds 1–7 were kept to prepare the references. Evaporation of the solvent from all solutions prepared and drying of the residue in vacuum desiccator was followed by its dissolving in CDCl3 (0.6 mL). 1H NMR spectra of chloroform solutions of the complexes were recorded within 1 h. The CIS values were obtained by subtraction of the δH7 and/or δH1′ values for the complexes from the chemical shift values of the neat compounds (reference). The NMR titrations were done for the constant concentration of 1–4. Equivalents of the solid titrants added equal to: 0.5–50 for 5, 1–10 for 6, and 1–20 for 7. The titration was continued to obtain the Δδ(H7) smaller than 0.1 ppm upon addition of the next portion. The δH7 (probe) obtained are collected in SI (titration charts). The Benesi–Hildebrandt [59] equation was used to calculate the K assoc.
X-ray
Single crystals of 2 and 4 used in the X-ray crystallographic experiment were obtained by slow evaporation of the solvent (chloroform) from NMR tube. The structural data for these compounds were collected at 123 ± 2 K with a Bruker–Nonius KappaCCD diffractometer equipped with APEXII detector using the graphite monochromatized MoKα radiation (λ = 0.71073 Å). Data were processed with DENZO-SMN [60]. The structures were solved by direct methods, using SIR-2004 [61], and refined on F 2, using SHELXL-97 [61]. The reflections were corrected for Lorenz polarization effects and absorption correction was not used. The H atoms bonded to C atoms were calculated to their idealized positions with isotropic temperature factors (1.2 times the C atom temperature factor) and refined as riding atoms. The H atoms bonded to N atoms were found from electron density map and fixed to distances of 0.88 Å from N atom with isotropic temperature factor (1.2 times the N atom temperature factor). The figures were drawn with ORTEP-3 [54] and MERCURY [62]. Other experimental X-ray data are shown in Table 2. CCDC-763984 (2) and CCDC-763985 (4) contain the supplementary crystallographic data for this article. These data can be obtained free of charge at http://www.ccdc.cam.ac.uk/conts/retrieving.html [or from the Cambridge Crystallographic Data Centre (CCDC), 12, Union Road, Cambridge CB2 1EZ, UK; fax: +44 1223 336033].
Calculations
Calculations at the M05/6-31G(2d,p) level for geometry optimizations of all structures studied have been performed in Gaussian [63]. The energy minimum was confirmed by the frequency calculations (all positive frequencies were obtained). Energy of the complex formation was calculated as the difference between energy of the complex and a sum of the energies of its constituents. The basis set superposition error (BSSE) correction was used with default settings. The single-point calculations (MP2/6-31G(2d,p) level) in GAMESS [64] at the geometry taken from M05/6-31G(2d,p) optimizations provided the orbital contours that were drawn with the use of QMView [65].
References
Watson JD, Crick FHC (1953) Nature 171:737
Jeffrey GA, Saenger W (1994) Hydrogen bonding in biological structures, study edition. Springer, Berlin
Eldrup AB, Christensen C, Haaima G, Nielsen PE (2002) J Am Chem Soc 124:3254
Prins LJ, Reinhoudt DN, Timmerman P (2001) Angew Chem Int Ed 40:2382
Lehn J-M (2000) In: Ciferri A (ed) Supramolecular polymer chemistry—scope and perspectives. Marcel Dekker, New York
Brunsveld L, Folmer BJB, Meijer EW, Sijbesma RP (2001) Chem Rev 101:4071
De Greef TFA, Smulders MMJ, Wolffs M, Schenning APHJ, Sijbesma RP, Meijer EW (2009) Chem Rev 109:5687
Alvares-Rua C, Garcia-Granda S, Goswami S, Mukherjee R, Dey S, Claramunt RM, Santa Maria MD, Rozas I, Jagerovic N, Alkorta I, Elguero J (2004) New J Chem 28:700
Newman SG, Taylor A, Boyd RJ (2008) Chem Phys Lett 450:210
Pranata J, Wierschke SG, Jorgensen WL (1991) J Am Chem Soc 113:2810
Jorgensen WL, Pranata J (1990) J Am Chem Soc 112:2008
Taubitz J, Lüning U (2009) Aust J Chem 62:1550
Sijbesma RP and Meijer EW (2003) Chem. Commun. 5
Blight BA, Camara-Campos A, Djurdjevic S, Kaller M, Leigh DA, McMillan FM, McNab H, Slawin AMZ (2009) J Am Chem Soc 131:14116
Murray TJ, Zimmerman SC, Kolotuchin SV (1995) Tetrahedron 51:635
Bell DA, Anslyn EV (1995) Tetrahedron 51:7161
Djurdjevic S, Leigh DA, McNab H, Parsons S, Teobaldi G, Zerbetto F (2007) J Am Chem Soc 129:476
Hisamatsu Y, Fukumi Y, Shirai N, Ikeda S-i, Odashima K (2008) Tetrahedron Lett 49:2005
McGhee AM, Kilner C, Wilson AJ (2008) Chem Commun 344
Xu W, Li X-C, Tan H, Chen G-J (2006) Phys Chem Chem Phys 8:4427
Park TK, Schroeder J, Rebek J (1991) J Am Chem Soc 113:5125
Gooch A, McGhee AM, Renton LC, Plante JP, Lindsay CI, Wilson AJ (2009) Supramol Chem 21:12
Sontjens SHM, Sijbesma RP, van Genderen MHP, Meijer EW (2000) J Am Chem Soc 122:7487
Schneider H-J (2009) Angew Chem Int Ed 48:3924
Scherman OA, Ligthart GBWL, Sijbesma RP, Meijer EW (2006) Angew Chem Int Ed 45:2072
Brammer S, Lüning U, Kühl C (2002) Eur J Org Chem 2002:4054
Manesiotis P, Hall AJ, Sellergren B (2005) J Org Chem 70:2729
Brown HC (1945) J Am Chem Soc 67:378
Brown HC (1945) J Am Chem Soc 67:1452
Lüning U, Kühl C (1998) Tetrahedron Lett 39:5735
Luening U, Kuhl C, Uphoff A (2002) Eur J Org Chem 2002:4063
Benoit RL, Fréchette M, Lefebvre D (1988) Can J Chem 66:1159
Beijer FH, Sijbesma RP, Vekemans JAJM, Meijer EW, Kooijman H, Spek AL (1996) J Org Chem 61:6371
Llanes-Pallas A, Matena M, Jung T, Prato M, Stohr M, Bonifazi D (2008) Angew Chem Int Ed 47:7726
Li Z, Ding J, Robertson G, Day M, Tao Y (2005) Tetrahedron Lett 46:6499
Katsuhiro I (2010) Chem Commun 2947
Ikeda C, Nagahara N, Motegi E, Yoshioka N, Inoue H (1999) Chem Commun 1759
Liu Y, Li Y, Jiang L, Gan H, Liu H, Li Y, Zhuang J, Lu F, Zhu D (2004) J Org Chem 69:9049
Liu Y, Zhuang J, Liu H, Li Y, Lu F, Gan H, Jiu T, Wang N, He X, Zhu D (2004) Chem Phys Chem 5:1210
Schmidt J, Schmidt R, Wurthner F (2008) J Org Chem 73:6355
Kang SO, Day VW, Bowman-James K (2009) J Org Chem 75:277
Lo PK, Sleiman HF (2008) Macromolecules 41:5590
Xu Z, Daka P, Wang H (2009) Chem Commun 6825
Chattopadhyay P, Pandey PS (2007) Bioorg Med Chem Lett 17:1553
Bolz I, Moon C, Enkelmann V, Brunklaus G, Spange S (2008) J Org Chem 73:4783
Forbes CC, Beatty AM, Smith BD (2001) Org Lett 3:3595
Feibush B, Figueroa A, Charles R, Onan KD, Feibush P, Karger BL (1986) J Am Chem Soc 108:3310
Breinlinger E, Niemz A, Rotello VM (1995) J Am Chem Soc 117:5379
Bolz I, Spange S (2008) Chem Eur J 14:9338
Chattopadhyay P, Pandey PS (2006) Tetrahedron 62:8620
Murray TJ, Zimmerman SC (1992) J Am Chem Soc 114:4010
Fenlon EE, Murray TJ, Baloga MH, Zimmerman SC (1993) J Org Chem 58:6625
Leung M-K, Mandal AB, Wang C-C, Lee G-H, Peng S-M, Cheng H-L, Her G-R, Chao I, Lu H-F, Sun Y-C, Shiao M-Y, Chou P-T (2002) J Am Chem Soc 124:4287
Farrugia LJ (1997) J Appl Crystallogr 30:565
Osmialowski B (2009) J Mol Struct THEOCHEM 908:92
Bernstein J, Stearns B, Shaw E, Lott WA (1947) J Am Chem Soc 69:1151
Kurita K, Williams RL (1975) J Heterocycl Chem 12:789
Kolehmainen E, Ośmiałowski B, Krygowski TM, Kauppinen R, Nissinen M, Gawinecki R (2000) J Chem Soc Perkin Trans 2:1259
Benesi H, Hildebrand J (1949) J Am Chem Soc 71:2703
Otwinowski Z, Minor W (1997) Methods in enzymology. In: Carter CW, Sweet RM (eds) Macromolecular crystallography, vol 276. Academic Press, New York
Burla MC, Camalli M, Carrozzini B, Cascarano GL, Giacovazzo C, Polidori G, Spagna R (2005) J Appl Crystallogr 38:381
Macrae CF, Bruno IJ, Chisholm JA, Edgington PR, Pidcock E, McCabe P, Rodriguez-Monge L, Taylor R, van de Streek J, Wood PA (2008) J Appl Crystallogr 41:466
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery J, J. A., Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian 03, Revision E.01. Gaussian Inc., Pittsburgh
Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su SJ, Windus TL, Dupuis M, Montgomery JA (1993) J Comput Chem 14:1347
Baldridge KK, Greenberg JP (1995) J Mol Graph 13:63
Acknowledgements
Financial support from the Polish Ministry of Science and Higher Education (Grant no. NN204 174138) is gratefully acknowledged. The authors are very much indebted to the Academic Computer Centre in Gdansk–TASK and CYFRONET in Cracow for providing computer time and programs.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ośmiałowski, B., Kolehmainen, E., Gawinecki, R. et al. NMR and quantum chemical studies on association of 2,6-bis(acylamino)pyridines with selected imides and 2,2′-dipyridylamine. Struct Chem 21, 1061–1067 (2010). https://doi.org/10.1007/s11224-010-9646-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-010-9646-2