Abstract
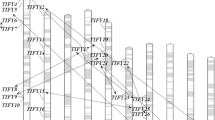
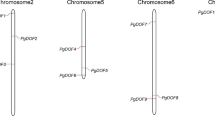
Deoxyribonucleic acid binding with one finger (Dof) family, a kind of plant-specific transcription factor family, is involved in a variety of biological processes including signal transduction, morphogenesis, and environmental stress responses. In the present study, 46 putative Dof genes were identified from maize (Zea mays subsp. mays). The predicted ZmDof genes were non-randomly distributed within their chromosomes, and segmental duplication seemed to be the prevalent mechanism for their expansion. Most of these genes lacked introns and some only possessed one intron. Here, we classified 140 Dof proteins from Arabidopsis, rice, sorghum, and maize into seven groups by means of phylogenetic analysis. In addition, many cis-elements responding to light, endosperm specific gene expression, hormone, meristem specific expression, and stress were also found in the 1,000 bps upstream sequences of promoter regions. Selection analysis also identified some significant site-specific constraints acted on most Dof paralogs. Expression profiles based on microarray data provided insights into the functional divergence of different tissues. Results by quantitative reverse transcription polymerase chain reaction analysis indicated that some ZmDofs responded to some abiotic stresses. Taken together, our study will provide some useful information for biological evolution and functional characterization of the maize Dof genes.






Similar content being viewed by others
Abbreviations
- Dof:
-
DNA binding with one finger
- NJ:
-
neighbor joining
- MEME:
-
multiple EM for motif elicitation
- TSS:
-
transcription start site
References
Baumann K, De Paolis A, Costantino P, Gualberti G (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11(3):323–334
Blanc G, Wolfe KH (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16(7):1667–1678
Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J, Zhang J, Wang T, Li H, Ye Z (2013) Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol 55(6):552–566
Cao J (2012) The pectin lyases in Arabidopsis thaliana: evolution, selection and expression profiles. PLoS ONE 7:e46944
Cao J, Shi F (2012) Dynamics of arginase gene evolution in metazoans. J Biomol Struct Dyn 30:407–418
Cao J, Shi F, Liu X, Huang G, Zhou M (2010) Phylogenetic analysis and evolution of aromatic amino acid hydroxylase. FEBS Lett 584:4775–4782
Cao J, Huang J, Yang Y, Hu X (2011) Analyses of the oligopeptide transporter gene family in poplar and grape. BMC Genomics 12:465
Chen Y, Cao J (2014) Comparative genomic analysis of the Sm gene family in rice and maize. Gene 539:238–249
Chen W, Chao G, Singh KB (1996) The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J 10:955–966
Chen Y, Hao X, Cao J (2014) Small auxin up-regulated RNA (SAUR) gene family in maize: identification, evolution and its phylogenetic comparison with Arabidopsis, rice and sorghum. J Integr Plant Biol 56(2):133–150
Chou PY, Fasman GD (1974) Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry 13(2):211–222
Comeron JM (1999) K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15:763–764
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14(6):1188–1190
Dong G, Ni Z, Yao Y, Nie X, Sun Q (2007) Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol Biol 63(1):73–84
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5(5):199–206
Farré D, Albà MM (2010) Heterogeneous patterns of gene-expression diversification in mammalian gene duplicates. Mol Biol Evol 27(2):325–335
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42(Database issue):D222–230
Gaut BS, Doebley JF (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94:6809–6814
Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93:10274–10279
Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2002) Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14:1253–1263
Guo Y, Qiu LJ (2013) Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 8(9):e76809
Hernando-Amado S, González-Calle V, Carbonero P, Barrero-Sicilia C (2012) The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol 12:202
Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11(11):550–558
Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 Fbox protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309:293–297
Jiang HY, Luo C, Jiang T, Cheng BJ, Zhu S (2010) Genome-wide analysis of Dof transcription factor family genes in maize. China J Bioinforma 8:198–201, abstract in English
Kim HS, Kim SJ, Abbasi N, Bressan RA, Yun DJ, Yoo SD, Kwon SY, Choi SB (2010) The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J 64:524–535
Kisu Y, Harada Y, Goto M, Esaka M (1997) Cloning of the pumpkin ascorbate oxidase gene and analysis of a cis-acting region involved in induction by auxin. Plant Cell Physiol 38:631–637
Kisu Y, Ono T, Shimofurutani N, Suzuki M, Esaka M (1998) Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant Cell Physiol 39:1054–1064
Kushwaha H, Gupta S, Singh VK, Rastogi S, Yadav D (2011) Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol Biol Rep 38:5037–5053
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3:17
Liu Y, Jiang H, Chen W, Qian Y, Ma Q, Cheng B, Zhu S (2011) Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul 63:225–234
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25(4):402–408
Martin C, Paz-Ares J (1997) MYB transcription factors in plants. Trends Genet 13(2):67–73
Marzábal P, Gas E, Fontanet P, Vicente-Carbajosa J, Torrent M, Ludevid MD (2008) The maize Dof protein PBF activates transcription of gamma-zein during maize seed development. Plant Mol Biol 67(5):441–54
Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P (1998) An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J 16(1):53–62
Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P (2002) A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol 130:111–119
Moreno-Risueno MA, Martínez M, Vicente-Carbajosa J, Carbonero P (2007) The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genomics 277(4):379–390
Negi J, Moriwaki K, Konishi M, Yokoyama R, Nakano T, Kusumi K, Hashimoto-Sugimoto M, Schroeder JI, Nishitani K, Yanagisawa S, Iba K (2013) A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr Biol 23:479–484
Noguero M, Atif RM, Ochatt S, Thompson RD (2013) The role of the DNA-binding one zinc finger (DOF) transcription factor family in plants. Plant Sci 209:32–45
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10(2):79–87
Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2000) Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev 14:28–33
Papi M, Sabatini S, Altamura MM, Hennig L, Schafer E, Costantino P, Vittorioso P (2002) Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol 128:411–417
Plesch G, Ehrhardt T, Mueller-Roeber B (2001) Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J 28:455–464
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66(4):553–563
Shaw LM, McIntyre CL, Gresshoff PM, Xue GP (2009) Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genomic 9(4):485–498
Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwaśniewski M, Zanor MI, Lohmann JU, De Veylder L, Witt I, Mueller-Roeber B (2008) The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J 56:779–792
Soderlund C, Bomhoff M, Nelson WM (2011) SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res 39:e68
Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T (2007) Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res 35(Web Server issue):W506–511
Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18:207–208
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends Plant Sci 12(6):239–44
Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94(14):7685–7690
Washio K (2001) Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim Biophys Acta 1520:54–62
Wasserman WW, Sandelin A (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5:276–287
Yanagisawa S (1997) Dof DNA binding domains of plant transcription factors contribute to multiple protein–protein interactions. Eur J Biochem 250(2):403–410
Yanagisawa S (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21:281–288
Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45(4):386–91
Yanagisawa S, Izui K (1993) Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem 268(21):16028–16036
Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17(2):209–214
Yanagisawa S, Sheen J (1998) Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10:75–89
Yang X, Tuskan GA, Cheng MZ (2006) Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol 142(3):820–830
Yang Z, Wang X, Gu S, Hu Z, Xu H, Xu C (2008) Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407(1–2):1–11
Yu CS, Lin CJ, Hwang JK (2004) Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 13:1402–1406
Zhang B, Chen W, Foley RC, Buttner M, Singh KB (1995) Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell 7:2241–2252
Zhu X, Ma H, Chen Z (2011) Phylogenetics and evolution of Su(var)3-9 SET genes in land plants: rapid diversification in structure and function. BMC Evol Biol 11:63
Acknowledgments
This project is supported by grants from the National Science Foundation of China (No. 31100923), the National Science Foundation of Jiangsu Province (BK2011467), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Jiangsu University “Youth Backbone Teacher Training Project” (2012–2016) to JC.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Data S1
Alignment data used in this study. (TXT 129 kb)
Fig. S1
Neighbor-joining phylogenetic tree of Dof proteins in Arabidopsis, rice, sorghum, and maize. (PDF 61 kb)
Fig. S2
Distribution of the predicted positive selection sites on the secondary structure of Dof protein in group VII. The predicted positive selection sites are indicated with arrows and red bold letters. (PDF 56 kb)
Table S1
Distribution of five class’s cis-elements in the promoter regions of the 46 ZmDof genes. (XLS 39 kb)
Table S2
Primers used in this study. (DOC 31 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Cao, J. Comparative Analysis of Dof Transcription Factor Family in Maize. Plant Mol Biol Rep 33, 1245–1258 (2015). https://doi.org/10.1007/s11105-014-0835-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0835-9