Abstract
Background and aims
Microorganisms colonize plant roots for mutual benefits. Colonization is initiated by the soil microbial community but is also affected by soil conditions and plant type. Rice typically grows under wetland conditions that are anoxic, thus being supportive for an anaerobic methanogenic microbial community. Maize, however, grows under upland conditions that are oxic, thus being supportive for an aerobic microbial community. Crop rotation between wetland rice and upland maize is not uncommon, but the effect of this management on microbial colonization of plant roots is largely unknown and was the aim of our study.
Methods
We used the roots of rice and maize from a two-year study in the Philippines, where on the same soil wetland rice was cultivated either in both wet and dry season or was rotated with upland maize in the dry season. The microbial colonization of the root ecto- and endorhizosphere was assessed by using quantitative PCR and illumina sequencing of the bacterial and archaeal 16S rRNA genes.
Results
The data showed that maize roots had completely different microbial community structures than the rice roots from continuous wetland cultivation, while rice roots from crop rotation were in-between. These effects of management were seen for each of the different bacterial phyla. For example, among the most abundant operational taxonomic units (OTUs) Firmicutes, Deltaproteobacteria and the methanogenic Methanocella spp. were less abundant while Alphaproteobacteria and the methanogenic Methanobacterium spp. were more abundant on maize than on rice roots.
Conclusions
Our study showed that root colonization by Archaea and Bacteria was strongly affected by crop rotation between wetland rice and upland maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant roots are closely associated with microorganisms. On the average 17% of plant photosynthates are released to the soil environment and may serve as growth substrates for microorganisms (Jones et al. 2009). On the other hand, microorganisms benefit plants in multiple ways, e.g., by providing nutrients, protecting against pathogens, helping against stress etc. (Bais et al. 2006; Mendes et al. 2011; Yang et al. 2009). Studies on Arabidopsis thaliana, a model plant, indicated that the genotype of the plant had a measurable, albeit small effect on the root endophytes (Bulgarelli et al. 2012; Lundberg et al. 2012). Similar results were obtained with maize (Peiffer et al. 2013) and rice (Edwards et al. 2015). However, these studies also showed that the composition of the root microbial communities was strongly affected by the soil source, geographical location, and cultivation practice. Furthermore, composition depends on how the root microbial community is assessed, which basically is the extraction protocol. Thus, the root microbial community can be differentiated from inside to outside as endophytic (inside the root tissue), ectophytic (on the rhizoplane, i.e., the ectorhizosphere), and rhizospheric (soil particles firmly attached to the root surface) (Edwards et al. 2015; Hernandez et al. 2015).
Rice is a wetland plant grown in flooded soil. Flooding impedes the transport of oxygen, so that the bulk of the soil becomes anoxic and supportive for anaerobic degradation of organic matter and CH4 production (Liesack et al. 2000). However, the rice gas vascular system allows the transport of oxygen into the roots and the rhizosphere thus providing a habitat for both aerobic and anaerobic bacteria involved in redox reactions of the C, N and S cycle (Liesack et al. 2000; Sessitsch et al. 2012). Maize, on the other hand, is a crop grown in aerated upland soil, which is usually oxic and supportive for aerobic degradation of organic matter to CO2.
Crop rotation between flooded rice and a non-flooded maize is becoming increasingly popular, as it saves irrigation water and provides a cash crop (maize) in addition to the food crop (rice) (Timisina et al. 2010). Previous studies on rice-planted flooded soil showed that the microbial community in rhizospheric soil was different from that of unplanted bulk soil (Breidenbach et al. 2016b), while the difference between the microbial communities in rhizospheric soil from flooded rice and non-flooded maize was relatively small (Breidenbach and Conrad 2015). However, extended crop rotation showed that the archaeal community composition was affected if flooded rice was rotated with non-flooded maize. The bacterial community, on the other hand, was comparatively little affected (Breidenbach et al. 2016a).
In order to focus more definitely on the microbial community colonizing the plant roots, we used samples from the crop rotation experiment described by Breidenbach et al. (2016a) and extracted the microbial community from the ecto- and endorhizosphere. The crop rotation system was on a soil at the International Rice Research Institute (IRRI) in the Philippines, and consisted of triplicate fields that were grown for two years with flooded rice during both the wet and the dry season, and of triplicate fields that were grown for two years with flooded rice during the wet season and non-flooded maize during the dry season. In particular, we asked the questions (1) whether rice and maize roots supported different microbial communities, and (2) whether the colonization of rice roots was affected by the intermittent crop rotation.
Materials and methods
Sampling site and sample processing
The sampling site was located at the International Rice Research Institute (IRRI) in Los Banos, Philippines. Detailed site description can be found in Heinz et al. (2014). This work was part of the interdisciplinary project “Introducing Non-Flooded Crops in Rice-Dominated Landscapes: Impact on Carbon, Nitrogen, and Water Cycles (ICON)”. A detailed description of the field experiment can be found in Weller et al. (2015). The experiment consisted of a flooded rice–maize crop rotation (MR) and a control with only flooded rice (RR). Briefly, we studied fields cultivated with either irrigated rice or upland maize at the reproductive growth phase of the plants over three years in both the dry season and the wet season. Fields under crop rotation (MR) were drained and managed as upland fields cultivating upland maize (variety: Pioneer P3482YR) in the dry season and flooded again and cultivated with rice (irrigated lowland variety: NSIC Rc222) in the wet season. The control fields (RR) fields were flooded and cropped with rice both in the dry and the wet season with drainage in between. A detailed overview of the crop rotation system and the sampling time points is given in Breidenbach et al. (2016a). Fields were operated in triplicates (RR: fields No. 3,6,9; MR: fields No. 4,7,10) and managed with conventional N-fertilization (rice: seeding 30 kg N/ha, 30 kg P2O5/ha, 30 kg K2O/ha; at 28 and 55 days after seeding 50 kg N/ha; maize: 30 kg N/ha, 50 kg P2O5/ha, 3 0 kg K2O/ha; at 27–29 and 47–50 DAS 50 kg N/ha). In each of these fields we randomly selected three sampling plots of one square meter and sampled one soil core (5 cm diameter, 20 cm length) from each plot.
Soil cores were always taken in the vicinity of a plant (ca. 10 cm). The soil contained numerous fine roots and thus was most probably influenced by the plant roots. However, no attempts were made to separate a specific rhizospheric soil compartment. Subsequently, soil samples of 5 g were taken from the middle of the core (~ 10 cm depth), added to 10 mL RNAlater© solution (Life Technologies, Darmstadt, Germany), kept on ice and later stored at −20 °C.
Within the sampled square meter one plant, rice respectively maize, was sampled. Plants were harvested and the roots were cut. After 3-times washing with tap water to remove adhering rhizospheric soil, the roots were fragmentized in small ~1-cm pieces. Subsequently, those were added to 10 mL RNAlater© solution (Life Technologies, Darmstadt, Germany), kept on ice and later stored at −20 °C.
DNA from soil and roots was extracted using the NucleoSpin® Soil Kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions. Before, cells were mechanically disrupted by bead-beating. Hence, the root extract contained microbes from both the ecto- and the endorhizosphere.
Quantitative polymerase chain reaction
The quantification of archaeal and bacterial 16S rDNA and of several functional marker genes was conducted using quantitative polymerase chain reaction (qPCR) based on a SYBRGreen approach. The primer combinations used were: Ba519f / Ba907r (Stubner 2002) for bacterial genes; Ar364f (Burggraf et al. 1997)/Ar934br (Grosskopf et al. 1998) for archaeal genes; MCRf/MCRr (Springer et al. 1995) for methanogens; A189-mb661 (Costello and Lidstrom, 1999) for methanotrophs; Arch-amoAF/Arch-amoAR (Francis et al. 2005) for archaeal ammonium oxidizers; and amoA-1F/amoA2R (Rotthauwe et al. 1997) for bacterial ammonium oxidizers. The protocols were used as described in the original publications.
Illumina amplicon sequencing
Illumina HiSeq2000 amplicon sequencing of the bacterial and archaeal community was conducted using primer combinations F515/R806 (Bates et al. 2011) targeting the V4 region of the 16S rRNA gene (approximately 250 nucleotides) for both archaea and bacteria. The forward primers were tagged with a unique 6-base pair barcode (list of barcodes see Table S1). Sequencing of the PCR products was done at the Max Planck Genome Centre (MPGC) in Cologne, Germany. Trimming forward and reverse adaptors from the merged sequences was done using the mothur software platform (Schloss et al. 2009). Sequence quality management and operational taxonomic units (OTU) analysis was conducted using UPARSE pipeline as described by Edgar (2013). Only microbial high-quality sequences with a minimum read length of 200 bp were used. Sequences that did not match the primer sequences and were smaller than 200 bp or contained any ambiguities were excluded from further analysis. Chimeric sequences were removed using UCHIME (Edgar et al. 2011). After denoising, sequences were aligned against the RDP database 16S rRNA gene database (Cole et al. 2014) using the naïve Bayesian classifier (Schloss et al. 2011; Wang et al. 2007). Sequences, which could not be assigned to bacteria or archaea, were discarded. OTUs were defined using a distance matrix with 3% dissimilarity (Zinger et al. 2011). Data analysis was performed using mothur software package version 1.31.2 (http://www.mothur.org/) following the standard operational procedure (SOP) (Schloss et al. 2009). The sequences were deposited at NCBI under the biosample accession numbers SAMN06462397 (forward sequences) and SAMN06462398 (reverse sequences).
Statistical analysis
Statistical analyses were conducted in R version 2.14.1 (R Development Core Team 2011). Analysis of variance (ANOVA) followed by Tukey’s post hoc test, non-metric multidimensional scaling (NMDS), canonical correspondence analysis (CCA) and PERMANOVA (ADONIS) were done with package vegan version 2.0.5 (Oksanen et al. 2012). The similarity indices used for NMDS and CCA were based on Bray-Curtis and Euklidian distances, respectively.
Results
Microbial abundance
The abundances of gene copy numbers on rice and maize roots were quantified in rice-maize (MR) crop rotation and in rice only (RR) control during the dry and wet seasons of 2012 and 2013 (Fig. 1). The field plots were adjacent on the same soil as described before (Breidenbach et al. 2016a; Weller et al. 2015). Significant differences in gene copy numbers between fields and plants are summarized in Table S2. Copy numbers of archaeal and bacterial 16S rRNA genes (Fig. 1a, b) on roots were different between rice and maize during the entire observational period. Archaeal numbers on maize roots were generally lower than on rice roots irrespectively of whether rice was grown on RR or MR fields, while numbers on rice roots were the same. The archaeal 16SrRNA gene abundance pattern was mirrored by the copy numbers of the mcrA gene coding for a subunit of the methyl coenzyme M reductase of methanogenic archaea (Fig. 1c). The mcrA copy numbers were on a range of 105 to 109 per gram dry root.
Copy numbers of different genes on roots of rice and maize plants grown under crop rotation (MR, grey bars) or as flooded rice only (RR, black bars) in 2012 and 2013 during the wet and the dry season. The following genes were determined using qPCR: (a) archaeal 16S rRNA gene; (b) bacterial 16S rRNA gene; (c) mcrA coding for a subunit of the methyl Coenzyme M reductase; (d) pmoA coding for a subunit of the particulate methane monooxygenase; (e) archaeal amoA coding for a subunit of the ammonium monooxygenase of Archaea; (f) bacterial amoA coding for a subunit of the ammonium monooxygenase of Bacteria. Asterics indicate significant (p < 0.05) differences in pairwise comparison between RR and MR. For further statistical evaluation see Table S2
Copy numbers were also determined for pmoA and amoA coding for subunits of the bacterial particulate methane monooxygenase (Fig. 1d) and the archaeal (Fig. 1e) and bacterial (Fig. 1f) ammonium monooxygenases, respectively. Copy numbers of pmoA were higher on rice roots than on maize roots, and the same was found for bacterial amoA, while copy numbers of archaeal amoA were similar across all treatments (Table S2).
Archaeal community composition
The archaeal community composition on plant roots was determined by illumina sequencing of the 16S rRNA gene (Fig. S1). Woesearchaeota, Euryarchaeota, Thaumarchaeota and Aenigmarchaeota showed the highest relative abundances among the archaea that could be classified. Among the methanogenic Euryarchaeota the genera with the highest number of different OTUs were Methanobacterium (4 OTUs), Methanospirillum (3 OTUs), Methanocella and Misancellaneous Euryarchaeota Group (MEG; 2 OTUs each), and Methanoregula, Methanosaeta and Methanosarcina (1 OTU each) (Fig. S1). The OTUs of Methanobacterium were the most abundant ones. Abundance on rice roots in RR stayed fairly constant from 2012 to 2013, while it was larger on maize roots in RM but decreased with time (Fig. 2). The abundance of the other methanogenic genera was generally lower. Methanocella was more abundant on rice roots than on maize roots, but the patterns of the other genera did not show a consistent change (Fig. 2).
OTU-based relative abundance of Archaea on roots of rice and maize plants grown under crop rotation (MR) or as flooded rice only (RR) in 2012 and 2013 during the wet and the dry season. The top 50 OTUs, based on relative abundance, were grouped according to their phylogenetic assignment on the level of (a) phyla and (b) genera. The bubble size indicates the relative abundance (1–30%), the phylogenetic groups were ordered according to the number of OTUs they represent
Overall, however, NMDS analysis showed that the archaeal communities were significantly different between rice roots in RR versus maize roots in MR, while those on rice roots in MR were in-between (Fig. 3a). This result was confirmed by CCA analysis, which considered not only the archaeal community composition, but also the effects of water supply, year of cultivation and plant type (Fig. S2a). The water supply was determined from data of rain and irrigation water listed in the study of Weller et al. (2016), which was simultaneously conducted within the ICON project on the same fields. The CCA indicated that the differences in community composition were affected by the different water supply in rice and maize fields. Considering all samples, archaeal and euryarchaeotal communities were significantly different (p < 0.01) comparing RR versus MR, and rice versus maize plants (Table S3), but the R-value was relatively low (0.32 to 0.43) (Table S3), except the correlation between total archaeal community and plant type (R = 0.72). Comparing only the communities on rice roots, field type (RR versus MR) showed the largest significant correlation (about R = 0.35) followed by sampling year (2012 versus 2013; about R = 0.23), while season (wet versus dry) had no effect (Table S4).
NMDS analysis of total (a) archaeal and (b) bacterial 16S rRNA gene OTUs detected on maize roots grown under crop rotation (MRmaize, open symbols) and on rice roots either under crop rotation (MRrice, grey symbols) or under flooded rice only (RR, closed symbols). For further explanation of the symbols see Fig. 5
Bacterial community composition
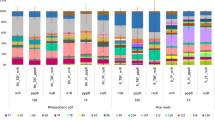
The bacterial community composition on plant roots was also determined by illumina sequencing of the 16S rRNA gene (Fig. 4). Proteobacteria was with 40–60% relative abundance the most abundant bacterial phylum, followed by Cyanobacteria, which made up to 40% relative abundance on maize roots but less than 10% on rice roots. Other conspicuous bacterial phyla were Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Planctomycetes, and Verrucomicrobia (Fig. 4). Beta-, Delta-, Alpha-, and Gammaproteobacteria were detected with 15, 13, 9 and 5 OTUs, respectively. Those of Gammaproteobacteria were generally the less abundant ones, those of Deltaproteobacteria were much less on maize than on rice roots (Fig. S3). Firmicutes and Cyanobacteria occurred with 6 and 5 OTUs, respectively. While the OTUs of Firmicutes were generally lowest on maize roots, those of Cyanobacteria were generally the highest (Fig. S3). The relative abundance of OTUs belonging to putatively aerobic methanotrophs (Proteobacteria) was generally less than 1%, the genus Methylocystis (<0.1–0.9%) being the most abundant one.
Overall, similarly as for Archaea, NMDS analysis showed that the bacterial communities were significantly different between rice roots in RR versus maize roots in MR, while those on rice roots in RM were in-between (Fig. 3b). CCA analysis again indicated that water supply to the fields was important for these differences (Fig. S2b). The same differentiation was observed in NMDS analyses of the individual bacterial phyla (Fig. 5), and also of putatively methanotrophic bacteria (Fig. S4). All 15 phyla analyzed showed a significant (p < 0.01) differentiation between rice and maize roots and between RR and MR plants. R-values were on the average R = 0.82 when comparing rice and maize roots and R = 0.35 when comparing RR and MR fields (Table S3). Comparing only the bacterial communities on rice roots, field type had only a significant effect on Chloroflexi (R = 0.43), while sampling year had effects on a number of different bacterial phyla, especially on Actinobacteria (R = 0.77), Bacteroidetes (R = 0.62), Chloroflexi and Armatimonadetes (both R = 0.5) (Table S4).
NMDS analysis of 16S rRNA gene OTUs assigned to major bacterial phyla detected on maize roots grown under crop rotation (MRmaize, open symbols) and on rice roots either under crop rotation (MRrice, grey symbols) or under flooded rice only (RR, closed symbols). The individual symbols indicate the field (RR, MR), the year (2012, 2013) and the season (D-dry, W-wet) when samples were taken
Interestingly, field type had not only a significant effect on the bacterial community on the roots but also on that in the soil, while plant type (in contrast to the community on root) had no significant effect on the soil bacterial community as shown by NMDS and CCA analysis (Fig. S5).
Discussion
Our study showed that rice roots and maize roots exhibited significantly different community structures of Archaea and Bacteria despite the fact that the plants were grown in the same soil. The difference was apparent both in abundance and composition. Even when assessing individual bacterial phyla, composition of the communities were different on rice and maize roots. The bacterial and archaeal community compositions grown on roots of rice that was rotated with maize were only slightly different from those of non-rotated rice. They actually were in-between non-rotated rice and maize, showing that it was not only the root environment of a particular plant, but also the management by crop rotation that affected root colonization, and presumably also differences in nutrient cycling. Since crop rotation between rice and maize was also a rotation between flooded soil and upland soil conditions, it is likely the water content and the aeration of the soil that affected root colonization. Indeed, the soil bacterial community in rotational fields was significantly different of that vegetated with non-rotated rice confirming previous observations (Breidenbach et al. 2016a).
Root associated microbiomes of rice are differentiated between root compartments (rhizosphere, rhizoplane or ectorhizosphere, endorhizosphere), field sites (locations across California’s Central valley), cultivation practice (EcoFarm, Organic), and rice variety (six cultivars) indicating that colonization of the roots is affected by the soil microbiota and the plant genotype (Edwards et al. 2015). Soil environment and plant genotype also shape the microbiota of the rhizosphere of maize, although soil environment seems to be the most crucial factor, as samples across different locations in the USA have a more pronounced effect than samples of 27 different maize genotypes (Peiffer et al. 2013). Nevertheless, plant genotype is an essential factor as demonstrated for rhizosphere bacterial communities in maize (Bouffaud et al. 2012), methanogenic archaeal communities in rice (Conrad et al. 2008), and methanotrophic bacterial communities in rice (Lüke et al. 2011). The present study extends these results to two different plant species, maize and rice, which exhibited completely different ecto- and endorhizosphere communities although they were grown in the same soil.
However, the soil type is not the only factor shaping the soil microbial community. In fact, local soil environmental conditions, water status in particular, are very important as seen by the effects of flooding on the development of the soil microbial community (Lueders and Friedrich 2000; Noll et al. 2005) and their transcription activity (Kim and Liesack 2015; Ma et al. 2012; Shrestha et al. 2009). These studies show that the soil microbial community changes because of adaptation from aerobic to anaerobic conditions. Therefore, it is mainly the different soil microbial community in flooded versus upland soil that results in different microbial communities on the rhizoplane. Effects of water management on rhizoplane communities have been observed in poplar trees (Graff and Conrad 2005) and are also seen here in rice-maize crop rotation. While the rhizoplane community of rice roots was adapted to the flooded conditions, that of maize roots was adapted to upland conditions.
Rice roots are colonized by a community of anaerobic bacteria and methanogenic archaea that are able to produce CH4, most probably not only from soil organic matter but also from root exudates derived from photosynthetically fixed carbon (Pump and Conrad 2014; Watanabe et al. 1999; Yuan et al. 2012). Maize fields, however, that are cultivated under drained upland conditions do not show production and emission of CH4 as seen on the experimental fields in our study (Kraus et al. 2015; Weller et al. 2015). Although both maize as well as rice roots contained archaeal OTUs from all major methanogenic taxonomic orders, the absolute abundance of methanogenic archaea was lower on maize than on rice roots. For Bacteria, on the other hand, absolute abundance was similar for rice and maize roots. However, composition of bacterial communities were different. In general, roots were dominated by the phylum Proteobacteria, similarly as observed in other studies with different plant roots (Bulgarelli et al. 2012; Edwards et al. 2015; Lundberg et al. 2012; Peiffer et al. 2013). However, while rice roots were dominated by OTUs from the Deltaproteobacteria, maize roots were dominated by Alphaproteobacteria. Deltaproteobacteria typically comprise bacteria with anaerobic metabolism (often obligatorily), such as iron reducers, sulfate reducers and syntrophic fermenting bacteria. Among the Alphaproteobacteria it is apparently the obligately aerobic genus Burkholderia that colonizes maize roots (Bouffaud et al. 2012; Peiffer et al. 2013), and also the aerobic methanotrophs, such as Methylocystis. However, methanotrophs were not among the 100 most common OTUs. Firmicutes comprising Clostridia as fermenting bacteria were mainly found on rice roots, while they were less abundant on maize roots, again emphasizing the fact that rice roots in contrast to maize roots develop in an anoxic environment, where fermentation and methanogenesis occurs. However, since the bacterial OTUs were all based on 16S rRNA gene phylogeny, analysis of microbial functions is limited to common knowledge of the physiology of microbial taxa.
In conclusion, the results of our study answered our initial questions and showed that the microbiomes of maize and rice roots were different and that crop rotation between rice and maize affected the community structures on rice roots. However, our study also indicated that much of the differences between the microbiomes on rice versus maize root was due to the different soil environmental conditions, i.e. anoxic flooded soil supporting anaerobic microbial metabolism on rice roots versus oxic drained soil supporting aerobic microbial metabolism on maize roots.
References
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interations with plants and other organisms. Annu Rev Plant Biol 57:233–266
Bates ST, Cropsey GWG, Caporaso JG, Knight R, Fierer N (2011) Bacterial communities associated with the lichen symbiosis. Appl Environ Microbiol 77:1309–1314
Bouffaud ML, Kyselkova M, Gouesnard B, Grundmann G, Muller D, Moenne-Loccoz Y (2012) Is diversification history of maize influencing selection of soil bacteria by roots? Mol Ecol 21:195–206
Breidenbach B, Conrad R (2015) Seasonal dynamics of bacterial and archaeal methanogenic communities in flooded rice fields and effect of drainage. Front Microbiol 5:752. doi:10.3389/fmicb.2014.00752
Breidenbach B, Blaser MB, Klose M, Conrad R (2016a) Crop rotation of flooded rice with upland maize impacts the resident and active methanogenic microbial community. Environ Microbiol 18:2868–2885
Breidenbach B, Pump J, Dumont MG (2016b) Microbial community structure in the rhizosphere of rice plants. Front Microbiol 6:1537. doi:10.3389/fmicb.2015.01537
Bulgarelli D, Rott M, Schlaeppi K, VerLorenVanThemaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95
Burggraf S, Huber H, Stetter KO (1997) Reclassification of the crenarchaeal orders and families in accordance with 16S rRNA sequence data. Int J Syst Bacteriol 47:657–660
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal database project: data and tools for high troughput rRNA analysis. Nucleic Acids Res 42:D633–D642
Conrad R, Klose M, Noll M, Kemnitz D, Bodelier PLE (2008) Soil type links microbial colonization of rice roots to methane emission. Glob Chang Biol 14:657–669
Costello AM, Lidstrom ME (1999) Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl.Environ.Microbiol. 65:5066–5074
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2220
Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688
Graff A, Conrad R (2005) Impact of flooding on soil bacterial communities associated with poplar (Populus sp.) trees. FEMS Microbiol Ecol 53:401–415
Grosskopf R, Janssen PH, Liesack W (1998) Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol 64:960–969
Heinz E, Kraft P, Buchen C, Frede HG, Aquino E, Breuer L (2014) Set up of an automatic water quality sampling system in irrigated agriculture. Sensors 14:212–228
Hernandez M, Dumont MG, Yuan Q, Conrad R (2015) Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl Environ Microbiol 81:2244–2253
Jones DL, Nguyen C, Finlay RD (2009) Carbon flow in the rhizosphere: carbon trading at the soil-root interface [review]. Plant Soil 321:5–33
Kim Y, Liesack W (2015) Differential assemblage of functional units in paddy soil microbiomes. PLoS One 10:e0122221. doi:10.1371/journal.pone.0122221
Kraus D, Weller S, Klatt S, Haas E, Wassmann R, Kiese R, Butterbach-Bahl K (2015) A new landscape DNDC biogeochemical module to predict CH4 and N2O emissions from lowland rice and upland cropping systems. Plant Soil 386:125–149
Liesack W, Schnell S, Revsbech NP (2000) Microbiology of flooded rice paddies [review]. FEMS Microbiol Rev 24:625–645
Lueders T, Friedrich M (2000) Archaeal population dynamics during sequential reduction processes in rice field soil. Appl Environ Microbiol 66:2732–2742
Lüke C, Bodrossy L, Lupotto E, Frenzel P (2011) Methanotrophic bacteria associated to rice roots: the cultivar effect assessed by T-RFLP and microarray analysis. Environ Microbiol Rep 3:518–525
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, DelRio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90
Ma K, Conrad R, Lu Y (2012) Responses of methanogen mcrA genes and their transcripts to an alternate dry/wet cycle of paddy field soil. Appl Environ Microbiol 78:445–454
Mendes R, Kruijt M, DeBruijn I, Dekkers E, VanderVoort M, Schneider JH, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA, Raaijmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Noll M, Matthies D, Frenzel P, Derakshani M, Liesack W (2005) Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ Microbiol 7:382–395
Oksanen J, Blanchet GF, Kindt R, Legendre R, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MH, Szoecs E, and Wagner H (2012) Vegan: Community ecology package ver.2.0–2.5. available online at: http://cran.r-project.org/web/packages/vegan/index.html,
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548–6553
Pump J, Conrad R (2014) Rice biomass production and carbon cycling in 13CO2 pulse-labeled microcosms with different soils under submerged conditions. Plant Soil 384:213–229
R Development Core Team (2011) R: a language and environment for statistical computing. The R foundation for statistical computing, Vienna, Austria; http://www.R-project.org/,
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker - molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson C, Sahl JW, Stres B, Thallinger GG, VanHorn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi:10.1371/journal.pone.0027310
Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, Woyke T, Mitter B, Hauberg-Lotte L, Friedrich F, Rahalkar M, Hurek T, Sarkar A, Bodrossy L, VanOverbeek L, Brar D, VanElsas J, Reinhold-Hurek B (2012) Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant-Microbe Interactions 25:28–36
Shrestha PM, Kube M, Reinhardt R, Liesack W (2009) Transcriptional activity of paddy soil bacterial communities. Environ Microbiol 11:960–970
Springer E, Sachs MS, Woese CR, Boone DR (1995) Partial gene sequences for the a subunit of methyl-coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int J Syst Bacteriol 45:554–559
Stubner S (2002) Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen ™ detection. J Microbiol Methods 50:155–164
Timisina J, Jat ML, Majumdar K (2010) Rice-maize systems of South Asia: current status, future prospects and research priorities for nutrient management [review]. Plant Soil 335:65–68
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Watanabe A, Takeda T, Kimura M (1999) Evaluation of origins of CH4 carbon emitted from rice paddies. J Geophys Res 104:23623–23629
Weller S, Kraus D, Ayag KR, Wassmann R, Alberto M, Butterbach-Bahl K, Kiese R (2015) Methane and nitrous oxide emissions from rice and maize production in diversified rice cropping systems. Nutr Cycl Agroecosyst 101:37–53
Weller S, Janz B, Jörg L, Kraus D, Racela HS, Wassmann R, Butterbach-Bahl K, Kiese R (2016) Greenhouse gas emissions and global warming potential of traditional and diversified tropical rice rotation systems. Glob Chang Biol 22:432–448
Yang J, Kloepper JW, Ryu CM (2009) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14:1–4
Yuan Q, Pump J, Conrad R (2012) Partitioning of CH4 and CO2 production originating from rice straw, soil and root organic carbon in rice microcosms. PLoS One 7:e49073. doi:10.1371/journal.pone.0049073
Zinger L, Amaral-Zettler LA, Fuhrman JA, Horner-Devine M, Huse SM, Welch DB, Martiny JB, Sogin M, Boetius A, Ramette A (2011) Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One 6:e24570. doi:10.1371/journal.pone.0024570
Acknowledgements
Open access funding provided by Max Planck Society. This work has been funded as part of the ICON consortium (BR2238/9-1). We thank the German Research Foundation (DFG) for funding (FOR 1701, ‘Introducing Non-Flooded Crops in Rice-Dominated Landscapes: Impacts on Carbon, Nitrogen and Water Cycles [ICON]’. Furthermore, we thank the International Rice Research Institute and Reiner Wassmann for providing research space and support during sample collection. We thank Mary Louise Mendoza, Eugene Aquino and Jerico Stefan Bigornia for sample collection. We thank the Max-Planck-Genome-Center in Cologne for access to the sequencing facility and support. We also thank Hannes Schmidt and an anonymous reviewer for helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Sven Marhan.
Electronic supplementary material
ESM 1
(PDF 1061 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Breidenbach, B., Brenzinger, K., Brandt, F.B. et al. The effect of crop rotation between wetland rice and upland maize on the microbial communities associated with roots. Plant Soil 419, 435–445 (2017). https://doi.org/10.1007/s11104-017-3351-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3351-5