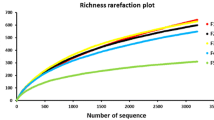
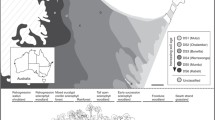
Abstract
Aims
We refine and test a next-generation sequencing assay for the molecular identification and quantification of plant roots from mixed-species samples.
Methods
We modified primers targeting the trnL intron to provide greater taxonomic resolution and developed an improved bioinformatics pipeline that can identify roots based on global, site-, and plot-specific taxon lists. We tested our primers and pipeline on mock plant communities of known composition and mixed-species samples of roots collected from temperate grassland and high Arctic tundra communities.
Results
We retrieved a high correlation (0.72) between observed and expected community dissimilarities. We found positive linear relationships between aboveground and belowground species richness in the grassland community, with higher correlations for plot-specific reference databases (R = 0.70) than a global reference database (R = 0.48). This highlights the importance of local reference databases within the bioinformatics pipeline. Lower root than aboveground richness suggests that typical root lateral spread in this grassland is less than 25 cm. We observed lateral rooting extents of up to 40 cm and rooting depths of up to 30 cm for six high Arctic species.
Conclusions
Testing on mock community, grassland, and tundra samples demonstrated that next-generation sequencing using our improved trnL primers and pipeline can successfully recover plant root community structure.



Similar content being viewed by others
References
Bast F, Rani P, Meena D (2014) Chloroplast DNA phylogeography of holy basil (Ocimum tenuiflorum) in Indian Subcontinent. Sci World J 2014:847482. doi:10.1155/2014/847482
Bliss LC, Henry GHR, Svoboda J, Bliss DI (1994) Patterns of plant distribution within two polar desert landscapes. Arct Alp Res 26:46. doi:10.2307/1551876
Bobowski BR, Hole D, Wolf PG, Bryant L (1999) Identification of roots of woody species using polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) analysis. Mol Ecol 8:485–491
Brunner I, Brodbeck S, BÜchler U, Sperisen C (2001) Molecular identification of fine roots of trees from the Alps: reliable and fast DNA extraction and PCR–RFLP analyses of plastid DNA. Mol Ecol 10:2079–2087. doi:10.1046/j.1365-294X.2001.01325.x
Buermans HPJ, den Dunnen JT (2014) Next generation sequencing technology: advances and applications. Biochim Biophys Acta (BBA) - Mol Basis Dis 1842:1932–1941. doi:10.1016/j.bbadis.2014.06.015
Cahill JF, McNickle GG (2011) The behavioral ecology of nutrient foraging by plants. Annu Rev Ecol Syst 42:289–311
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi:10.1093/bioinformatics/btp636
Cayuela L, Granzow-de la Cerda Í, Albuquerque FS, Golicher DJ (2012) Taxonstand: An r package for species names standardisation in vegetation databases. Methods Ecol Evol 3:1078–1083. doi:10.1111/j.2041-210X.2012.00232.x
Chapin FS, Bret-Harte MS, Hobbie SE, Zhong H (1996) Plant functional types as predictors of transient responses of arctic vegetation to global change. J Veg Sci 7:347–358
Coupland RT, Johnson RE (1965) Rooting characteristics of grassland species in Saskatchewan. J Ecol 53:475–507
Development Core Team R (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi:10.1093/bioinformatics/btq461
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381
Frank DA, Pontes AW, Maine EM, Caruana J, Raina R, Raina S, Fridley JD (2010) Grassland root communities: species distributions and how they are linked to aboveground abundance. Ecology 91:3201–3209. doi:10.1890/09-1831.1
Frank DA, Pontes AW, Maine EM, Fridley JD (2015) Fine-scale belowground species associations in temperate grassland. Mol Ecol 24:3206–3216. doi:10.1111/mec.13232
Haling R, Simpson R, McKay A, Hartley D, Lambers H, Ophel-Keller K, Wiebkin S, Herdina H, Riley I, Richardson A (2011) Direct measurement of roots in soil for single and mixed species using a quantitative DNA-based method. Plant Soil 348:123–137. doi:10.1007/s11104-011-0846-3
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic acids symposium series
Hiiesalu I, Öpik M, Metsis M, Lilje L, Davison J, Vasar M, Moora M, Zobel M, Wilson SD, Pärtel M (2012) Plant species richness belowground: higher richness and new patterns revealed by next-generation sequencing. Mol Ecol 21:2004–2016. doi:10.1111/j.1365-294X.2011.05390.x
Hiiesalu I, Pärtel M, Davison J, Gerhold P, Metsis M, Moora M, Öpik M, Vasar M, Zobel M, Wilson SD (2014) Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass. New Phytol 203:233–244. doi:10.1111/nph.12765
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143. doi:10.1186/gb-2007-8-7-r143
Iversen CM, Sloan VL, Sullivan PF, Euskirchen ES, McGuire AD, Norby RJ, Walker AP, Warren JM, Wullschleger SD (2015) The unseen iceberg: plant roots in arctic tundra. New Phytol 205:34–58. doi:10.1111/nph.13003
Jackson RB, Canadell J, Ehleringer JR, Mooney HA, Sala OE, Schultze ED (1996) A global analysis of root distributions for terrestrial biomes. Oecologia 108:389–411
Kesanakurti PR, Fazekas AJ, Burgess KS, Percy DM, Newmaster SG, Graham SW, Barrett SCH, Hajibabaei M, Husband BC (2011) Spatial patterns of plant diversity below-ground as revealed by DNA barcoding. Mol Ecol 20:1289–1302. doi:10.1111/j.1365-294X.2010.04989.x
Klimeš L (2003) Scale-dependent variation in visual estimates of grassland plant cover. J Veg Sci 14:815–821. doi:10.1111/j.1654-1103.2003.tb02214.x
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. doi:10.1093/bioinformatics/btm091
Kowalczyk R, Taberlet P, Coissac E, Valentini A, Miquel C, Kamiński T, Wójcik JM (2011) Influence of management practices on large herbivore diet—case of European bison in Białowieża Primeval Forest (Poland). For Ecol Manag 261:821–828. doi:10.1016/j.foreco.2010.11.026
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi:10.1128/aem.01043-13
Linder CR, Moore LA, Jackson RB (2000) A universal molecular method for identifying underground plant parts to species. Mol Ecol 9:1549–1559. doi:10.1046/j.1365-294x.2000.01034.x
McNickle GG, Cahill JF, Deyholos MK (2008) A PCR-based method for the identification of the roots of 10 co-occurring grassland species in mesocosm experiments. Botany 86:485–490
Moore LA, Field CB (2005) A technique for identifying the roots of different species in mixed samples using nuclear ribosomal DNA. J Veg Sci 16:131–134
Newmaster SG, Fazekas AJ, Ragupathy S (2006) DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Can J Bot 84:335–341. doi:10.1139/b06-047
Nguyen NH, Smith D, Peay K, Kennedy P (2015) Parsing ecological signal from noise in next generation amplicon sequencing. New Phytol 205:1389–1393. doi:10.1111/nph.12923
Oksanen JF, Blanchet G, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2015) Vegan: community ecology package v.2.3–0
Pärtel M, Hiiesalu I, Öpik M, Wilson SD (2012) Below-ground plant species richness: new insights from DNA-based methods. Funct Ecol 26:775–782. doi:10.1111/j.1365-2435.2012.02004.x
Pegard A, Miquel C, Valentini A, Coissac E, Bouvier F, François D, Taberlet P, Engel E, Pompanon F (2009) Universal DNA-based methods for assessing the diet of grazing livestock and wildlife from feces. J Agric Food Chem 57:5700–5706. doi:10.1021/jf803680c
Phillips M (2011) The depth distribution of organic carbon in mineral cryosols at two sites in the Canadian Arctic. University of Saskatchewan, Saskatoon, Soil Science
Piper C, Lamb E, Siciliano S (2015a) Smooth brome changes gross soil nitrogen cycling processes during invasion of a rough fescue grassland. Plant Ecol 216:235–246. doi:10.1007/s11258-014-0431-y
Piper CL, Siciliano SD, Winsley T, Lamb EG (2015b) Smooth brome invasion increases rare soil bacterial species prevalence, bacterial species richness and evenness. J Ecol 103:386–396. doi:10.1111/1365-2745.12356
Plant Working Group CBOL, Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim K-J, Kress WJ, Schneider H, van AlphenStahl J, Barrett SCH, van den Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacón J, Clark A, JJ C, Conrad F, DS D, CS F, TAJ H, ML H, BC H, LJ K, PR K, JS K, Y-D K, Lahaye R, H-L L, DG L, Madriñán S, Maurin O, Meusnier I, SG N, C-W P, DM P, Petersen G, JE R, GA S, Savolainen V, Seberg O, MJ W, D-K Y, DP L (2009) A DNA barcode for land plants. Proc Natl Acad Sci U S A 106:12794–12797. doi:10.1073/pnas.0905845106
Pompanon F, Deagle BE, Symondson WOC, Brown DS, Jarman SN, Taberlet P (2012) Who is eating what: diet assessment using next generation sequencing. Mol Ecol 21:1931–1950. doi:10.1111/j.1365-294X.2011.05403.x
Price JN, Hiiesalu I, Gerhold P, Pärtel M (2012) Small-scale grassland assembly patterns differ above and below the soil surface. Ecology 93:1290–1296. doi:10.1890/11-1942.1
Pucheta E, Bonamici I, Cabido M, Diaz S (2004) Below-ground biomass and productivity of a grazed site and a neighbouring ungrazed exclosure in a grassland in Central Argentina. Austral Ecol 29:201–208
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. doi:10.1186/1471-2105-12-38
Rayé G, Miquel C, Coissac E, Redjadj C, Loison A, Taberlet P (2010) New insights on diet variability revealed by DNA barcoding and high-throughput pyrosequencing: chamois diet in autumn as a case study. Ecol Res 26:265–276. doi:10.1007/s11284-010-0780-5
Ridgway KP, Duck JM, Young JPW (2003) Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnl (UAA) intron. BMC Ecol 3:8
Schenk HJ, Jackson RB (2002) Rooting depths, lateral root spreads and below-ground/above-ground allometries of plants in water-limited ecosystems. J Ecol 90:480–494
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/AEM.01541-09
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS one 6:e27310. doi:10.1371/journal.pone.0027310
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res 35:e14. doi:10.1093/nar/gkl938
Taggart JM, Cahill JF Jr, McNickle GG, Hall JC (2011) Molecular identification of roots from a grassland community using size differences in fluorescently labelled PCR amplicons of three cpDNA regions. Mol Ecol Resour 11:185–195. doi:10.1111/j.1755-0998.2010.02893.x
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tuomisto H (2012) An updated consumer’s guide to evenness and related indices. Oikos 121:1203–1218. doi:10.1111/j.1600-0706.2011.19897.x
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. doi:10.1093/nar/gks596
Vittoz P, Bayfield N, Brooker R, Elston DA, Duff EI, Theurillat J-P, Guisan A (2010) Reproducibility of species lists, visual cover estimates and frequency methods for recording high-mountain vegetation. J Veg Sci 21:1035–1047. doi:10.1111/j.1654-1103.2010.01216.x
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new Bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi:10.1128/AEM.00062-07
Acknowledgments
This work was funded by NSERC Discovery grants to SDS and EGL. Drs. Ben Raymond and John McKinlay (Australian Antarctic Division) and Dr. Shaun Neilsen (University of New South Wales, Sydney Australia) provided expertise in development of R code for the pipeline automation stages. The manuscript was substantially improved based on comments from Katherine Stewart and several anonymous reviewers. The authors declare no conflict of interest with this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Peter J. Gregory.
Electronic supplementary material
ESM 1
(DOCX 1.22 mb)
Rights and permissions
About this article
Cite this article
Lamb, E.G., Winsley, T., Piper, C.L. et al. A high-throughput belowground plant diversity assay using next-generation sequencing of the trnL intron. Plant Soil 404, 361–372 (2016). https://doi.org/10.1007/s11104-016-2852-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-016-2852-y