Abstract
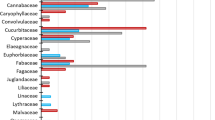
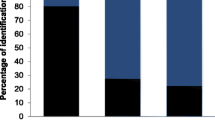
Characterizing the diet of large herbivores and the determinants of its variation remains a difficult task in wild species. DNA-based techniques have the potential to complement traditional time-consuming methods based on the microhistology of plant cuticle fragments in fecal or rumen samples. Recently, it has been shown that a short chloroplast DNA fragment, the P6 loop of the trnL (UAA) intron, can act as a minimalist barcode. Here, we used the trnL approach with high-throughput pyrosequencing to study diet from feces in a wild herbivore, the alpine chamois (Rupicapra rupicapra) and showed that the fine resolution in plant determination obtained with this method allows exploring subtle temporal shifts and inter-individual variability in diet composition. First, we built a DNA barcoding database of 475 plants species. Seventy-two percent of plant species can be unambiguously identified to species level, 79% to genus level and 100% to family level using the P6 loop. Second, we analysed 74 feces collected from October to November. Based on 47,896 P6 loop sequences, we identified a total of 110 taxa, 96 in October and 76 in November, with a clear diet shift between October and November. We recognized four and two clusters of feces composition in October and November, respectively, revealing different diet categories among individuals within each month. DNA-based diet analysis is faster and more taxonomically precise than studies based on microhistology, and opens new possibilities for analysing plant-herbivore interactions in the wild.




Similar content being viewed by others
References
Anthony RG, Smith NS (1974) Comparison of rumen and fecal analysis to describe deer diets. J Wildl Manag 38:729–746
Auvray F (1983) Recherche sur l’éco-éthologie du mouflon (Ovis musimon musimon Schreber 1782) dans le massif de Caroux-Espinousse (Hérault). PhD thesis, Montpellier University, France
Babad G (1997) Etude des relations entre un peuplement animal et la végétation: impacts du chamois, du chevreuil et du mouflon sur les peuplements forestiers dans la Réserve Nationale de Faune Sauvage et de Chasse des Bauges (Savoie). PhD thesis, Chambery University, France
Berducou C (1974) Contribution à l’étude d’un problème éco-physiologique pyrénéen: l’alimentation hivernale de l’isard. PhD thesis, Toulouse University, France
Bertolino S, di Montezemolo NC, Bassano B (2009) Food-niche relationships within a guild of alpine ungulates including an introduced species. J Zool 277:63–69
Bjelland T, Ekman S (2005) Fungal diversity in rock beneath a crustose lichen as revealed by molecular markers. Microb Ecol 49:598–603
Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques 20:1008–1010
Calenge C, Darmon G, Basille M, Loison A, Jullien JM (2007) The factorial decomposition of the Mahalanobis distances in habitat selection studies. Ecology 89:555–566
Castle EJ (1956) The rate of passage of foodstuffs through the alimentary tract of the goat. 3. The intestines. Br J Nutr 10:338–346
Chapuis JL, Bousses P, Pisanu B, Reale D (2001) Comparative rumen and fecal diet microhistological determinations of European mouflon. J Range Manag 54:239–242
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Couturier MAJ (1938) Le Chamois. Arthaud, Grenoble
Crawley MJ (2007) The R book. Wiley, New York
Darmon G, Calenge C, Loison A, Maillard D, Jullien J-M (2007) Social and spatial patterns determine the population structure and colonisation processes in mouflon. Can J Zool 85:634–643
De Jong CB, Gill RMA, van Wieren SE, Burlton FWE (1995) Diet selection by roe deer Capreolus capreolus in Kielder forest in relation to plant cover. For Ecol Manag 79:91–97
Deagle BE, Tollit DJ, Jarman SN, Hindell MA, Trites AW, Gales NJ (2005) Molecular scatology as a tool to study diet: analysis of prey DNA in scats from captive Steller sea lions. Mol Ecol 14:1831–1842
Deagle BE, Eveson JP, Jarman SN (2006) Quantification of damage in DNA recovered from highly degraded samples—a case study on DNA in faeces. Front Zool 3:11
Deagle BE, Gales NJ, Evans K, Jarman SN, Robinson S, Trebilco R, Hindell MA (2007) Studying seabird diet through genetic analysis of faeces: a case study on macaroni penguins (Eudyptes chrysolophus). PLoS ONE 2:e831
Dixon P (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930
Dove H, Mayes RW (1996) Plant wax components: a new approach to estimating intake and diet composition in herbivores. J Nutr 126:13–26
Ferrari C, Rossi G, Cavani C (1988) Summer food-habits and quality of female, kid and subadult Apennine chamois, Rupicapra pyrenaica ornata Neumann, 1899 (Artiodactyla, Bovidae). J Mamm Biol 53(3):170–177
Foley WJ, McIlwee A, Lawler I, Aragones L, Wollnough AP, Berding N (1998) Ecological applications of near infrared reflectance spectroscopy—a tool for rapid, cost-effective prediction of the composition of plant and animal tissues and aspects of animal performance. Oecologia 116:293–305
Garcia Gonzalez R, Cuartas P (1995) Trophic utilization of a montane/subalpine forest by chamois (Rupicapra pyrenaica) in the Central Pyrenees. In: Conference on ungulates in temperate forest ecosystems, 23–27 April. Elsevier Science, Wageningen, pp 15–23
Garel M, Loison A, Jullien JM, Dubray D, Maillard D, Gaillard JM (2009) Sex-specific growth in mass in Alpine chamois. J Mammal 90:954–960
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp 41:95–98
Hofmann RR (1989) Evolutionary steps of ecophysiological adaptation and diversification of ruminants: a comparative view of their digestive system. Oecologia 78:443–457
Holechek JL, Vavra M, Pieper RD (1982) Botanical composition determination of range herbivore diets—a review. J Range Manag 35:309–315
Höss M, Kohn M, Pääbo S, Knauer F, Schröder W (1992) Excrement analysis by PCR. Nature 359:199
Howe HF (1988) Ecological relationships of plants and animals. Oxford University Press, Oxford
Kohn MH, Wayne RK (1997) Facts from faeces revisited. TREE 12:223–227
Kozena I (1986) Further data on the winter diet of the chamois, Rupicapra rupicapra rupicapra, in the Jeseniky. Folia Zool 35:207–214
La Morgia V, Bassano B (2009) Feeding habits, forage selection, and diet overlap in Alpine chamois (Rupicapra rupicapra L.) and domestic sheep. Ecol Res 24:1043–1050
Lembke M (2005) L’habitat alimentaire du bouquetin des Alpes (Capra i. ibex) au cours de la saison de végétation sur le massif de Belledonne—Sept Laux (Isère, France). Variations spatio-temporelles, effets du dimorphisme sexuel et implications pour sa gestion. PhD thesis, Chambery University, France
Loison A, Appolinaire J, Jullien JM (2006) How reliable are population counts to detect trends in population size in chamois? Wildl Biol 12:77–88
Loison A, Darmon G, Cassar S, Jullien J-M, Maillard D (2008) Age- and sex-specific settlement patterns of chamois (Rupicapra rupicapra) offspring. Can J Zool 86:588–593
Magnuson VL, Ally DS, Nylund SJ et al (1996) Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. BioTechniques 21:700–709
Noris JJ (1943) Botanical analyses of stomach contents as a method for determining forage consumption of range sheep. Ecology 24:244–251
Obrtel R, Holisova V, Kozena I (1984) The winter diet of the chamois Rupicapra rupicapra rupicapra, in the Jeseniky Mts. Folia Zool 33:327–338
Pegard A, Miquel C, Valentini A, Coissac E, Bouvier F, Francois D, Taberlet P, Engel E, Pompanon F (2009) Universal DNA-based methods for assessing the diet of grazing livestock and wildlife from feces. J Agric Food Chem 57:5700–5706
Poinar HN, Hofreiter M, Spaulding WG, Martin PS, Stankiewicz BA, Bland H, Evershed RP, Possnert G, Pääbo S (1998) Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 281:402–406
Shipley LA (2007) The influence of bite size on foraging at larger spatial and temporal scales by mammalian herbivores. Oïkos 116:1964–1974
Soininen EM, Valentini A, Coissac E, Miquel C, Gielly L, Brochmann C, Brysting AK, Sønstebø JH, Ims RA, Yoccoz NG, Taberlet P (2009) Analysing diet of small herbivores: the efficiency of DNA barcoding coupled with high-throughput pyrosequencing for deciphering the composition of complex plant mixture. Front Zool 6:16
Stewart DRM (1967) Analysis of plant epidermis in faeces: a feeding experiment with penned sika deer. J Mamm Soc Jpn 7:167–180
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL(UAA) intron for plant DNA barcoding. Nucleic Acid Res 35:e14
Team RDC (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Thioulouse J, Chessel D, Doledee S, Olivier JM (1997) ADE-4A multivariate analysis and graphical display software. Stat Comput 7:75–83
Valentini A, Miquel C, Nawaz MA, Bellemain E, Coissac E, Pompanon F, Gielly L, Cruaud C, Nascetti G, Wincker P et al (2009a) New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol Ecol Res 9:51–60
Valentini A, Pompanon F, Taberlet P (2009b) DNA barcoding for ecologists. Trends Ecol Evol 9:51–60
Vaucher CA (1988) Contribution à l’étude éco-éthologique du chamois (Rupicapra rupicapra) au Mont Salève (Haute Savoie). PhD thesis, Univ Nancy 1
Yockney IJ, Hickling GJ (2000) Distribution and diet of chamois (Rupicapra rupicapra) in Westland forests, South Island, New Zealand. N Z J Ecol 24:31–38
Acknowledgments
We thank Nicolas Valy and Bruno Gravelat for their contribution for plant sampling and determination. We also thank Delphine Rioux, Ludovic Gielly and Alice Valentini for their technical help in the laboratory, and David Christianson and Jessica Rocha for correcting the English.
Author information
Authors and Affiliations
Corresponding author
Additional information
G. Rayé and C. Miquel contributed equally to this work.
Appendices
Appendix 1
See Table 3.
Appendix 2
See Table 4.
About this article
Cite this article
Rayé, G., Miquel, C., Coissac, E. et al. New insights on diet variability revealed by DNA barcoding and high-throughput pyrosequencing: chamois diet in autumn as a case study. Ecol Res 26, 265–276 (2011). https://doi.org/10.1007/s11284-010-0780-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11284-010-0780-5