Abstract
Key message
The overexpression of RXam2, a cassava NLR (nucleotide-binding leucine-rich repeat) gene, by stable transformation and gene expression induction mediated by dTALEs, reduce cassava bacterial blight symptoms.
Abstract
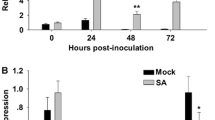
Cassava (Manihot esculenta) is a tropical root crop affected by different pathogens including Xanthomonas phaseoli pv. manihotis (Xpm), the causal agent of cassava bacterial blight (CBB). Previous studies have reported resistance to CBB as a quantitative and polygenic character. This study sought to validate the functional role of a NLR (nucleotide-binding leucine-rich repeat) associated with a QTL to Xpm strain CIO151 called RXam2. Transgenic cassava plants overexpressing RXam2 were generated and analyzed. Plants overexpressing RXam2 showed a reduction in bacterial growth to Xpm strains CIO151, 232 and 226. In addition, designer TALEs (dTALEs) were developed to specifically bind to the RXam2 promoter region. The Xpm strain transformed with dTALEs allowed the induction of the RXam2 gene expression after inoculation in cassava plants and was associated with a diminution in CBB symptoms. These findings suggest that RXam2 contributes to the understanding of the molecular basis of quantitative disease resistance.






Similar content being viewed by others
References
Allem A (1999) The closest wild relatives of cassava (Manihot esculenta Crantz). Euphytica 107:123–133
Alvarez E, Llano GA, Mejía JF (2012) Cassava diseases in Latin America, Africa and Asia. In: Howeler R (ed) The cassava handbook—a reference manual based on the asian regional cassava training course, held in Thailand. CIAT, Cali, pp 258–304
Bredeson JV, Lyons JB, Prochnik SE, Wu GA, Ha CM, Edsinger-Gonzales E, Grimwood J, Schmutz J, Rabbi IY, Egesi C et al (2016) Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nature Biotech 34:562–570
Chege MN, Wamunyokoli F, Kamau J, Nyaboga EN (2017) Phenotypic and genotypic diversity of Xanthomonas axonopodis pv. manihotis causing bacterial blight disease of cassava in Kenya. J Appl Biol Biotechnol 5:38–44
Cock JH (1982) Cassava: a basic energy source in the tropics. Science 218:755–762
Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18:1247–1256
Constantin EC, Cleenwerck I, Maes M, Baeyen S, Van Malderghem C, De Vos P et al (2016) Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol 65:792–806. https://doi.org/10.1111/ppa.12461
Debieu M, Huard-chauveau C, Genissel A, Roux F, Roby D (2016) Quantitative disease resistance to the bacterial pathogen Xanthomonas campestris involves an Arabidopsis immune receptor pair and a gene of unknown function. Mol Plant Pathol 17:510–520
Díaz Tatis PA, Herrera Corzo M, Ochoa Cabezas JC, Medina Cipagauta A, Prías MA, Verdier V, Chavarriaga Aguirre P, López Carrascal CE (2018) The overexpression of RXam1, a cassava gene coding for an RLK, confers disease resistance to Xanthomonas axonopodis pv. manihotis. Planta 247:1031–1042
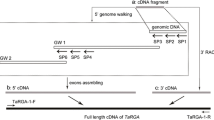
Diaz P (2016) Transference of RXam2 and Bs2 genes to confer resistance against cassava bacterial blight (CBB). PhD Dissertation. Universidad Nacional de Colombia
Duxbury Z, Ma Y, Furzer OJ, Huh SU, Cevik V, Jones JDG, Sarris PF (2016) Pathogen perception by NLRs in plants and animals: parallel worlds. BioEssays 38:769–781
French E, Kim B, Iyer-pascuzzi AS (2016) Mechanisms of quantitative disease resistance in plants. Semin Cell Dev Biol 56:201–208
Fukuoka S, Yamamoto S, Mizobuchi R, Yamanouchi U, Ono K, Kitazawa N, Yasuda N, Fujita Y, Thi T, Nguyen T et al (2014) Multiple functional polymorphisms in a single disease resistance gene in rice enhance durable resistance to blast. Sci Rep 4:1–7
Fukuoka S, Saka N, Mizukami Y, Koga H, Yamanouchi U, Yoshioka Y, Hayashi N, Ebana K, Mizobuchi R, Yano M (2015) Gene pyramiding enhances durable blast disease resistance in rice. Sci Rep 5:1–7
Gao Z, Chung E-H, Eitas TK, Dangl JL (2011) Plant intracellular innate immune receptor resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci USA 108:7619–7624
Geißler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J (2011) Transcriptional activators of human genes with programmable DNA-specificity. PLoS ONE 6:1–7
Gómez-Cano F, Soto J, Restrepo S, Bernal A, Lopez-Klein L, López C (2019) Gene co-expression network for Xanthomonas-challenged cassava reveals key regulatory elements of immunity processes. Eur J Plant Pathol 153:1083–1104. https://doi.org/10.1007/s10658-018-01628-4
Hahn SK, Terry ER, Leuschner K, Akobundu IO, Okali C, Lal R (1979) Cassava improvement in Africa. Field Crops Res 2:193–226
Hahn SK, Howland AK, Terry ER (1980) Correlated resistance of cassava to mosaic and bacterial blight diseases. Euphytica 29:305–311
Howles P, Lawrence G, Finnegan J, Mcfadden H, Ayliffe M, Dodds P (2005) Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol Plant Microbe Interact 18:570–582
Jorge V, Fregene MA, Duque MC, Bonierbale MW, Tohme J, Verdier V (2000) Genetic mapping of resistance to bacterial blight disease in cassava (Manihot esculenta Crantz). Theor Appl Genet 101:865–872
Jorge V, Fregene M, Velez CM, Duque MC, Tohme J, Verdier V (2001) QTL analysis of field resistance to Xanthomonas axonopodis pv. manihotis in cassava. Theor Appl Genet 102:564–571
Jubic LM, Saile S, Furzer OJ, El Kasmi F, Dangl JL (2019) Help wanted: helper NLRs and plant immune responses. Curr Opin Plant Biol 50:82–94
Kante M, Flores C, Moufid Y, Wonni I, Hutin M, Thomas E, Fabre S, Gagnevin L, Dagno K, Verdier V et al (2020) First report of Xanthomonas phaseoli pv. manihotis (Xpm), the causal agent of cassava bacterial blight (CBB) in Mali. Plant Dis 104:1852. https://doi.org/10.1094/PDIS-12-19-2611-PDN
Lolle S, Stevens D, Coaker G (2020) Plant NLR-triggered immunity: from receptor activation to downstream signaling. Curr Opin Immunol 62:99–105
López CE, Zuluaga AP, Cooke R, Delseny M, Tohme J, Verdier V (2003) Isolation of resistance gene candidates (RGCs) and characterization of an RGC cluster in cassava. Mol Genet Genomics 269:658–671
López CE, Quesada-Ocampo LM, Bohórquez A, Duque MC, Vargas J, Tohme J, Verdier V (2007) Mapping EST-derived SSRs and ESTs involved in resistance to bacterial blight in Manihot esculenta. Genome 50:1078–1088
Lozano J (1986) Cassava bacterial blight: a manageable disease. Plant Dis 70:1089–1093
Lozano R, Hamblin MT, Prochnik S, Jannink J-L (2015) Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics 16:360
Lukasik E, Takken FL (2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12:427–436
McCallum EJ, Anjanappa RB, Gruissem W (2017) Tackling agriculturally relevant diseases in the staple crop cassava (Manihot esculenta). Curr Opin Plant Biol 38:50–58
Michelmore RW, Christopoulou M, Caldwell KS (2013) Impacts of resistance gene genetics, function, and evolution on a durable future. Ann Rev Phytopathol 51:291–319
Monteiro F, Nishimura MT (2018) Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Ann Rev Phytopathol 56:243–267
Muñoz-Bodnar A, Perez-Quintero AL, Gomez-Cano F, Gil J, Michelmore R, Bernal A, Szurek B, Lopez C (2014) RNAseq analysis of cassava reveals similar plant responses upon infection with pathogenic and non-pathogenic strains of Xanthomonas axonopodis pv. manihotis. Plant Cell Rep 33:1901–1912. https://doi.org/10.1007/s00299-014-1667-7
Oldroyd GED, Staskawicz BJ (1998) Genetically engineered broad-spectrum disease resistance. Proc Natl Acad Sci USA 95:10300–10305
Olsen KM, Schaal B (1999) Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proc Natl Acad Sci USA 96:5586–5591
Pérez-Quintero AL, Rodriguez-R LM, Dereeper A, López C, Koebnik R, Szurek B, Cunnac S (2013) An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PLoS ONE 8:e68464
Pilet-Nayel ML, Moury B, Caffier V, Montarry J, Kerlan MC, Fournet S, Durel CE, Delourme R (2017) Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front Plant Sci 8:1838. https://doi.org/10.3389/fpls.2017.01838
Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 14:21–29
Qi D, DeYoung BJ, Innes RW (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158:1819–1832
Quang Huy N, Van QM, Quang Man L, Nguyen DT, Xuan Hoat T (2019) Identification of cassava bacterial blight-causing Xanthomonas axonopodis pv. manihotis based on rpoD and gyrB genes. Vietnam J Sci Technol 61:30–35
Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V (2013) Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet 9:e1003465
Roux F, Voisin D, Badet T, Balagu C, Barlet X, Huard-Chauveau C, Roby D, Raffaele S (2014) Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol Plant Pathol 15:427–432
Soto SJC, Mora MRE, Mathew B, Leon J, Gomez FA, Ballvora A, Lopez CCE (2017) Major novel QTL for resistance to Cassava bacterial blight identified through a multi-environmental analysis. Front Plant Sci 8:1169
St. Clair DA (2010) Quantitative disease resistance and quantitative resistance loci in breeding. Ann Rev Phytopathol 48:247–268
Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B (2013) Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol 200:808–819
Takahashi A, Hayashi N, Miyao A, Hirochika H (2010) Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging. BMC Plant Biol 10:175
Takken FLW, Goverse A (2012) How to build a pathogen detector: structural basis of NB-LRR function. Curr Opin Plant Biol 15:375–384
Takken FLW, Tameling WIL (2009) To nibble at plant resistance proteins. Science 324:744–746
Tao Y, Yuan FH, Leister RT, Ausubel FM, Katagiri F (2000) Mutational analysis of the Arabidopsis nucleotide binding site leucine-rich repeat resistance gene RPS2. Plant Cell 12:2541–2554
Taylor N, Gaitan-Solis E, Moll T, Trauterman B, Jones T et al (2012) A high-throughput platform for the production and analysis of Transgenic Cassava (Manihot esculenta) plants. Trop Plant Biol. https://doi.org/10.1007/s12042-
Taylor RK, Griffin RL, Jones LM, Pease B, Tsatsia F, Fanai C, Macfarlane B, Dale CJ, Davis RI (2017) First record of Xanthomonas axonopodis pv. manihotis in Solomon Islands. Australas Plant Dis Notes 12:49
Trujillo CA, Ochoa JC, Mideros MF, Restrepo S, López C, Bernal A (2014) A complex population structure of the cassava pathogen Xanthomonas axonopodis pv. manihotis in recent years in the caribbean region of Colombia. Microb Ecol 68:155–167
van Wersch S, Li X (2019) Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci 24:688–699
van Ooijen G, Mayr G, Kasiem MMA, Albrecht M, Cornelissen BJC, Takken FLW (2008) Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59:1383–1397
Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291:118–120
Acknowledgements
PAD was supported with a scholarship for graduate students from Universidad Nacional de Colombia. This work was partially funded by the Colombian Administrative Department of Science and Technology, Colciencias (contract N° 0315-2013). Thanks to the Universidad Antonio Nariño Editorial Office for support in the English editing of this manuscript. The authors are grateful to Valerie Verdier for assisting the beginning of this project, Camilo Dorado for his support on the statistical analysis, Fabio Gómez for his support on the molecular cloning of RXam2, Tran Tu for guidance in the construction of dTALEs and Didier Marin for the assistance in the greenhouse inoculation experiments. PD and CL want to dedicate this manuscript to Amelia and Emilio younger sister and brother of RXAM2. The authors declare no conflict of interest.
Author information
Authors and Affiliations
Contributions
PAD, AM and JCO developed cassava transgenic plants. PAD performed transgenic plants characterization experiments and dTALEs construction. PC and BS provided resources and orientation for transgenic plant development and dTALEs construction, respectively. MR and CR performed dTALEs inoculation assays. PAD and CEL conceived the study and wrote the manuscript. All the authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing or financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.


11103_2021_1211_MOESM3_ESM.xlsx
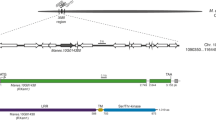
Supplementary file3 (XLSX 30 kb) Table S1 List of genes located near RXam2 (Manes.07G048100, 4853478-4857038) in a region that spans 4.8 Mb
Rights and permissions
About this article
Cite this article
Díaz-Tatis, P.A., Ochoa, J.C., Rico, E.M. et al. RXam2, a NLR from cassava (Manihot esculenta) contributes partially to the quantitative resistance to Xanthomonas phaseoli pv. manihotis. Plant Mol Biol 109, 313–324 (2022). https://doi.org/10.1007/s11103-021-01211-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01211-2