Abstract
Self-incompatibility (SI) is an intraspecific reproductive barrier that allows many families of flowering plants to prevent inbreeding and promote outcrosses. Extensive studies of SI in five families during the past more than two decades have revealed three distinct SI mechanisms. This chapter focuses on the mechanism employed by the Solanaceae, using mostly results obtained from Petunia. We first discuss the identification of two polymorphic genes at the S-locus, the S-RNase gene, which controls pistil specificity, and the S-locus F-box (SLF) gene, now named SLF1. For several years after its identification, SLF1 was thought to be solely responsible for pollen specificity, and biochemical models were developed based on this assumption. However, results inconsistent with this assumption were subsequently obtained, which led to the recent finding that pollen specificity is controlled by multiple, but an as yet unknown number of, polymorphic SLF genes located at the S-locus. A new model, named collaborative non-self recognition, has been proposed to explain the biochemical basis of specific inhibition of self-pollen tube growth. Based on this model, compatible pollination results from ubiquitination and subsequent degradation of non-self S-RNases collectively mediated by all SLF proteins, with each SLF responsible for detoxifying a subset of non-self S-RNases. We conclude this chapter with a discussion of some of the new questions raised by the finding that pollen specificity is controlled by multiple SLF genes.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In contrast to animals, plants cannot freely move about to select appropriate mates. To preserve species identity and to generate genetic diversity within species, it is imperative that plants possess mechanisms by which their female reproductive tissues, the pistils, can prevent unwanted or unsuitable pollen from delivering sperm cells to the ovary to effect fertilization. For example, many flowering plants that produce bisexual flowers have adopted a genetically controlled pre-zygotic barrier, called self-incompatibility (SI). SI involves a self/non-self recognition process between pollen and pistil. As a result of the recognition, self-pollen is rejected by the pistil to prevent inbreeding, and only non-self pollen is accepted to promote outcrosses. The specific recognition process involves the interaction between pistil specificity determinant and pollen specificity determinant, both of which are encoded by genes tightly linked at a highly polymorphic locus, named the S-locus. Variants of the S-locus are referred to as S-haplotypes and designated S 1, S 2, S 3, etc.
SI is quite common in flowering plants, but to date our understanding of the molecular and biochemical basis is limited to 5 of the estimated more than 60 families that possess SI. Extensive studies on these 5 families since the mid-1980s have revealed three distinct mechanisms (Takayama and Isogai 2005). In this chapter, we focus on the mechanism that is employed by the Solanaceae family (and likely by two other families, the Rosaceae and Plantaginaceae) using mostly the results obtained from the study of Petunia. We first describe the identification and characterization of the gene that controls pistil specificity, the S-RNase, and then describe the identification of the first S-locus F-box (SLF) gene, now named type-1 SLF (or SLF1), that is involved in controlling pollen specificity. After the identification of SLF1, it was thought to be the only gene that controls pollen specificity, as the prevailing thought then was that pollen specificity must be controlled by a single polymorphic gene, just like the prior finding that pistil specificity is controlled by a single polymorphic gene, S-RNase. Moreover, it is already difficult to envision how a single pollen specificity gene has coevolved with a single pistil specificity gene to maintain SI over the course of millions of years since this self/non-self recognition system was established. We thus next turn to the discussion of several lines of evidence, from the evolutionary perspective and experimental data, that are inconsistent with the assumption that a single SLF gene is solely responsible for pollen specificity, and describe the identification of additional SLF genes that are involved in pollen specificity. We present the latest model, named “collaborative non-self recognition,” that explains how the polymorphic S-RNase gene and multiple polymorphic SLF genes function in SI, allowing cross-compatible pollen tubes to grow through the pistil, but inhibiting the growth of self-pollen tubes in the pistil. Finally, we discuss some of the questions regarding the evolution, maintenance, and operation of S-RNase-based SI that were raised by the unexpected finding that pollen specificity is controlled by multiple SLF genes.
2 Identification and Characterization of the S-RNase Gene
To identify the gene that controls pistil specificity, pistil proteins that showed S-haplotype-specific differences in molecular mass and/or isoelectric point were first identified (Bredemeijer and Blaas 1981; Anderson et al. 1986). These proteins were deemed likely candidates, as allelic products of the pistil gene involved in SI were expected to be different in their amino-acid sequences such that they might be different in molecular mass or isoelectric point. This approach led to the identification of the S-RNase gene in Nicotiana alata (Anderson et al. 1986). However, the biochemical nature of S-RNase was not known until several years later when the amino-acid sequence of the fungal RNase T2 was determined (Kawata et al. 1988) and found to share a significant degree of sequence homology between its catalytic domain and a conserved domain of several allelic variants of S-RNase (McClure et al. 1989). Subsequently, S-RNase was shown to indeed possess RNase activity in vitro (McClure et al. 1989; Broothaerts et al. 1991; Singh et al. 1991). The S-RNase gene has been shown to be solely responsible for pistil specificity in SI by gain- and loss-of-function experiments (Lee et al. 1994; Murfett et al. 1994). For example, in Petunia inflata, expression of the S 3 -RNase gene in pistils of wild-type plants of S 1 S 2 genotype conferred on the pistils of the transgenic plants the ability to reject S 3 pollen, whereas expression of an antisense S 3 -RNase gene in wild-type plants of S 2 S 3 genotype led to the inability of the transgenic plants to reject S 3 pollen, but did not affect their ability to reject S 2 pollen (Lee et al. 1994). These experiments suggest that S-RNase is necessary and sufficient for the pistil to recognize and reject self-pollen.
The characteristics of S-RNase are also consistent with its involvement in pistil specificity. For example, S-RNase is specific to the pistil. It is first synthesized in the transmitting cell of the style and then secreted into the extracellular space of the transmitting tract. It is most abundant in the upper third segment of the style, where inhibition of incompatible pollen tubes occurs (Ai et al. 1990). Moreover, S-RNase has a high degree of allelic sequence diversity, with the highest divergent pair sharing only 38 % sequence identity (Tsai et al. 1992; McCubbin and Kao 2000), which is expected for a protein that is involved in self/non-self recognition. Sequence comparison revealed the presence of five conserved regions, C1–C5 (with C2 and C3 similar to the corresponding domains of RNase T2), and two hypervariable regions, HVa and HVb (Ioerger et al. 1991; Tsai et al. 1992; Takayama and Isogai 2005). Crystallographic analysis of a Solanaceae S-RNase showed that the protein-folding topology is typical for the RNase T2 family (Ida et al. 2001). Moreover, the results also showed that both HVa and HVb are exposed on the surface of the S-RNase protein and accessible to solvent, raising the possibility that these two regions have the allelic-specific function (Ida et al. 2001; Matsuura et al. 2001).
To further understand the function of S-RNase in SI, site-directed mutagenesis was used to replace the codon for one of the two catalytic His residues (located in C3) of S3-RNase of P. inflata with an Asn codon. The mutant S 3 -RNase gene was introduced into P. inflata plants of S 1 S 2 genotype, and the transgenic plants producing this mutated S3-RNase failed to reject S 3 pollen, suggesting that the RNase activity of S-RNase is an integral part of its function and that the biochemical mechanism of SI involves degradation of RNAs in incompatible pollen tubes (Huang et al. 1994). Moreover, as all S-RNases are glycoproteins with various numbers of N-linked glycan chains, a question was raised as to whether the recognition function of S-RNase resides in the glycan moiety, the protein backbone, or both. To address this question, the codon for the only potential N-glycosylation site of S3-RNase was replaced with a codon for Asp, and the mutant S 3 -RNase gene was introduced into wild-type plants of S 1 S 2 genotype to examine whether expression of the non-glycosylated S3-RNase could still confer on the transgenic plants the ability to reject S 3 pollen. The results showed that the non-glycosylated S3-RNase retained the full ability to reject self-pollen, suggesting that the recognition function of S-RNase resides in its amino-acid sequence (Karunanandaa et al. 1994). Domain-swapping experiments were performed to examine the role of the two hypervariable domains in allelic specificity (Kao and McCubbin 1996; Matton et al. 1997; Zurek et al. 1997; Matton et al. 1999). Swapping HVa and HVb between S11-RNase and S13-RNase of Solanum chacoense (which differ by four amino acids in the hypervariable regions) was sufficient to switch the allelic specificity (Matton et al. 1997, 1999). However, in the case of P. inflata and N. alata S-RNases examined, the two hypervariable regions are necessary but not sufficient for allele specificity. This conclusion is consistent with the finding that two P. inflata S-RNases, S6-RNase and S9-RNase, have identical sequences in HVa and differ by only two amino acids in HVb (Wang et al. 2001; Kao and Tsukamoto 2004).
For S-RNase to inhibit growth of self-pollen tubes, it must be able to enter incompatible pollen tubes to exert its cytotoxicity. Using immunocytochemistry, Luu et al. (2000) showed that S-RNase was localized in the cytoplasm of both self- and non-self pollen tubes. In contrast, Goldraij et al. (2006), using triple antibody-labeling immunolocalization, showed that S-RNase was initially sequestered in a vacuole-like compartment of both self- and non-self pollen tubes, but the compartment was later disrupted only in incompatible pollen tubes, releasing S-RNase into the cytoplasm. Both groups are in agreement that S-RNase is taken up by self- and non-self pollen tubes; however, the mechanism of S-RNase uptake is as yet unknown.
3 Identification of the First S-Locus F-box Gene, SLF1
One would expect that the approach used to successfully identify S-RNase and clone the S-RNase gene could be used to identify the pollen specificity determinant. However, no pollen protein that show S-haplotype specific differences in molecular mass or isoelectric point was ever identified. Another approach, RNA differential display, was used to identify pollen-specific genes that are tightly linked to the S-locus (Wang et al. 2003). This approach was based on the prediction that the gene controlling pollen specificity must be tightly linked to the S-RNase gene and shows allelic sequence diversity. Thirteen such genes of P. inflata were identified (Dowd et al. 2000; McCubbin et al. 2000). However, none of them was considered a good candidate for the pollen specificity gene because of their low allelic sequence diversity (Wang et al. 2003). Ultimately, it was through sequencing the S-locus region containing the S-RNase gene that a good candidate was first identified in Antirrhinum hispanicum (Plantaginaceae), which also possesses S-RNase-based SI (Lai et al. 2002). This gene is located ~9 kb downstream from the S 2 -RNase gene, and its deduced amino-acid sequence contains an F-box motif at the N-terminus. Thus, the gene was named AhSLF (A. hispanicum S-locus F-box gene). Subsequently, PmSLF, encoding an F-box protein, was identified in Prunus mume of Rosaceae through genomic sequence analysis of an ~60-kb region containing S-RNase (Entani et al. 2003), and SFB (S haplotype-specific F-box gene), also encoding an S-locus F-box protein, was identified in another rosaceous species, Prunus dulcis (almond) (Ushijima et al. 2003).
In P. inflata, PiSLF (P. inflata SLF) was identified from sequencing a 328-kb contig of an S-locus region containing the S 2 -RNase gene (Wang et al. 2004). PiSLF is located ~161 kb downstream from the S 2 -RNase gene. RNA blotting results showed that PiSLF is specifically expressed in developing pollen, mature pollen and pollen tubes. It also shows S-haplotype-specific restriction-fragment length polymorphism, as expected of a gene located at the S-locus. The deduced amino-acid sequences of three alleles (S 1 , S 2 , and S 3 ) of PiSLF show 10.3–11.6 % allelic sequence diversity. These properties of PiSLF suggest that it is a good candidate for the pollen specificity gene. PiSLF control of pollen specificity was established by a transgenic functional assay of its S 2 -allele, PiSLF 2 (Fig. 24.1a), designed based on the phenomenon named competitive interaction. This phenomenon was first observed in solanaceous species, including P. inflata and P. hybrida (Stout and Chandler 1942; Brewbaker and Natarajan 1960; Entani et al. 1999), where it was found that SI breaks down in heteroallelic pollen carrying two different S-alleles. For example, S 1 S 2 pollen produced by a tetraploid S 1 S 1 S 2 S 2 plant derived from self-incompatible plant S 1 S 2 loses its SI function and cannot be rejected by the S 1 S 1 S 2 S 2 pistil. Heteroallelic pollen can also result from duplication of one of the S-loci of self-incompatible plants carrying two different S-haplotypes (Golz et al. 1999). It was reasoned that if PiSLF controls pollen specificity, introducing PiSLF 2 into S 1 S 1 , S 1 S 2 , and S 2 S 3 plants should cause breakdown of SI in S 1 and S 3 pollen carrying the transgene, as a result of competitive interaction between the endogenous S 1 -allele or S 3 -allele of PiSLF and the introduced S 2 -allele. The results of these experiments were precisely as expected (Sijacic et al. 2004). For example (as shown in Fig. 24.1a), expression of a PiSLF 2 transgene in an S 2 S 3 transgenic plant caused breakdown of SI in S 3 pollen (heteroallelic), but not in S 2 pollen (homoallelic).
Transgenic functional assay showing that PiSLF is not the sole pollen specificity gene. (a) Self-pollination of an S 2 S 3 transgenic plant carrying a PiSLF 2 transgene. The transgenic plant produces four different genotypes of pollen as indicated. White triangles and blue triangles denote S2-RNase and S3-RNase molecules, respectively. Based on the findings that S 2 S 3 and S 3 S 3 , but no S 2 S 2 , genotypes were present in the self-progeny and that all progeny inherited the transgene (either one copy or two copies), only S 3 pollen carrying the PiSLF 2 transgene was accepted by the pistil. White and blue triangles marked with a broken line across denote those S2-RNases and S3-RNases, respectively, that are inside pollen tubes but fail to exert their cytotoxicity. That PiSLF 2 causes breakdown of SI in S 3 , but not in S 2 , pollen is consistent with the prediction by competitive interaction that SI breaks down in pollen carrying two different S-alleles. (b) Self-pollination of an S 2 S 3 transgenic plant carrying a PiSLF 3 transgene. The transgenic plant produces four different genotypes of pollen as indicated. White triangles and blue triangles denote S2-RNase and S3-RNase molecules, respectively. The transgenic plant remained self-incompatible, suggesting that all four genotypes of pollen were rejected by the pistil. White and blue triangles marked with a broken line across denote those S2-RNases and S3-RNases, respectively, that are inside pollen tubes but fail to exert their cytotoxicity. The finding that PiSLF 3 does not cause breakdown of SI in S 2 pollen is not consistent with the prediction by competitive interaction. (c) A protein degradation model based on the assumption that PiSLF is solely responsible for pollen specificity predicts that a PiSLF interacts with all S-RNases, except its self-S-RNase, to mediate their ubiquitination and ultimate degradation by the 26S proteasome. In the example shown, PiSLF2 interacts with S3-RNase and Sx-RNase (with Sx denoting S-RNases produced by any additional haplotype except S 2 ) to mediate their ubiquitination and degradation, as indicated by a broken line across S3-RNase and Sx-RNase; however, PiSLF2 does not interact with S2-RNase, its self-S-RNase. This model can explain the results of the transgenic functional assay shown in a. In S 3 pollen tubes expressing the PiSLF 2 transgene, PiSLF3 produced by the endogenous gene interacts with S2-RNase, and PiSLF2 produced from the transgene interacts with S3-RNase. As a result, both S2-RNase and S3-RNase are ubiquitinated and degraded, thus allowing the pollen tube to circumvent the toxic effect of these S-RNases. (d) Interpretation of the results of the transgenic functional assay shown in b based on the protein-degradation model shown in c. The failure of the PiSLF 3 transgene to cause breakdown of SI in S 2 pollen suggests that PiSLF3 cannot interact with S2-RNase, a non-self S-RNase, raising a question of whether PiSLF3 can interact with any other non-self S-RNases. Most importantly, this finding suggests that there must be additional protein(s) involved in pollen specificity that allow(s) S 3 pollen to interact with and detoxify S2-RNase to result in compatible pollination between S 3 pollen and S 2 -carrying pistils
As already stated, both self- and non-self S-RNases are taken up by a pollen tube, but only self-S-RNase can exert its cytotoxicity to inhibit the growth of the pollen tube. Before the discovery of the involvement of multiple SLF genes in pollen specificity (see Sect. 24.5 below), our lab proposed a protein degradation model, based on the assumption that PiSLF (SLF1) is the sole pollen determinant, to explain why only self-S-RNase can function inside a pollen tube (Hua and Kao 2006; Hua et al. 2008). The model predicts that for pollen of a given S-haplotype, (1) the allelic variant of PiSLF produced interacts with all its non-self S-RNases in the cytoplasm of the pollen tubes to mediate their ubiquitination and ultimate degradation by the 26S proteasome, thus allowing non-self pollen to effect fertilization, and (2) the allelic variant of PiSLF does not interact with its self-S-RNase, allowing the self-S-RNase to degrade pollen tube RNAs to cause growth inhibition of self-pollen tubes. These predictions were largely based on the findings by Hua and Kao (2006) that bacterially expressed S-RNases were ubiquitinated and degraded via the 26S proteasomal pathway in an in vitro cell-free system, and that an allelic variant of PiSLF interacted with its non-self S-RNases more strongly than with its self-S-RNase in an in vitro protein binding assay. As most F-box proteins are components of SCF complexes, a class of E3 ubiquitin ligase involved in ubiquitin-mediated protein degradation, the protein degradation model provided a reasonable explanation for the biochemical basis of the SI process. Moreover, the following findings are consistent with this model. (1) The subcellular localization of AhSLF-S2 (the S 2 -allelic variant of an SLF in Antirrhinum) detected by immunocytochemistry suggests that it is localized in the cytoplasm of pollen tubes (Wang and Xue 2005). (2) In S. chacoense, the level of S-RNase in compatible pollen tubes was ~30 % lower than that in incompatible pollen tubes (Liu et al. 2009). (3) Six lysine residues in S3-RNase of P. inflata were found to be necessary for targeting ubiquitination and degradation (Hua and Kao 2008).
This protein degradation model can explain why expression of a PiSLF 2 transgene in heteroallelic pollen causes breakdown of SI. For example, when a PiSLF 2 transgene is introduced into an S 2 S 3 plant, it causes specific breakdown of SI in S 3 transgenic pollen (Fig. 24.1a). As shown in Fig. 24.1c, in the S 3 transgenic pollen, PiSLF2 produced from the transgene interacts with S3-RNase, a non-self S-RNase, to mediate its ubiquitination and degradation. As PiSLF3 produced by the endogenous gene interacts with S2-RNase, a non-self S-RNase, to mediate its ubiquitination and degradation, both S2-RNase and S3-RNase taken up by the transgenic S 3 pollen tube are detoxified, allowing the S 3 transgenic pollen tube to be compatible with the S 3 -carrying pistil.
4 Evidence Against SLF1 Being Solely Responsible for Controlling Pollen Specificity
After the transgenic functional assay showing that one allele of PiSLF, PiSLF 2 , behaved as precisely as expected for the gene controlling pollen specificity, it was thought that the long search for the pollen specificity gene was over. However, further analyses of this gene, now named SLF1, revealed puzzling results that called into question this notion.
4.1 Evolutionary Consideration
First, the degree of sequence diversity of SLF1 is much lower than that of the S-RNase gene. For example, the allelic sequence diversity is only ~10 % for three alleles (S 1 , S 2, and S 3 ) of PiSLF, but ranges from 19 % to 26 % for the same three alleles of S-RNase. Most puzzlingly, the allelic sequence diversity of four alleles (S 1 , S 2 , S 4 , S 5 ) of AhSLF (A. hispanicum) ranges from only 1 % to 3 %, whereas that of the same four alleles of S-RNase ranges from 36 % to 51 % (Zhou et al. 2003; Newbigin et al. 2008). The high degree of allelic sequence diversity observed for S-RNase is what would be expected for a gene that controls pistil specificity in SI interactions with pollen. During the evolution of the SI system, any new S-haplotype that arises in a population has a reproductive advantage over all existing haplotypes, because its frequency is lower to begin with and thus has a higher probability of successful fertilization. As such, the new S-haplotype will increase in its frequency in the population. Under this kind of frequency-dependent selection, there is pressure for sequence divergence in the genes that constitute S-haplotype specificity to generate new S-haplotypes. Thus, the low degree of allelic sequence diversity of the SLF1 gene is not consistent with its presumed pollen specificity role in SI.
Second, the evolutionary history of S-RNase is consistent with the ancient origin of SI, but SLF1 seemed to have evolved much more recently. Phylogenetic studies of S-RNase alleles from several solanaceous species revealed that polymorphism of S-RNase predated divergence of these species (e.g., some P. inflata alleles are more similar to some N. alata alleles than to other P. inflata alleles) (Ioerger et al. 1990). Allelic polymorphism of S-RNase was shown to exist in the common ancestor of the solanaceous species 30–40 million years ago (Paape et al. 2008). However, phylogenetic studies of SLF1 alleles from P. inflata and A. hispanicum showed that SLF1 has a much shorter evolutionary history (Newbigin et al. 2008).
Third, the evolution of S-RNase and SLF1 seems not to be concordant. Coevolution of genes controlling pistil and pollen specificity has long been a central theory in SI. In Brassicaceae-type SI, the genes encoding pollen and style specificity determinants have been shown to be coevolved (Sato et al. 2002; Takebayashi et al. 2003). However, as stated here, S-RNase and SLF1 seem to have different evolutionary histories, and moreover, genealogies of S-RNase and SLF1 do not show a pattern of coevolution (Newbigin et al. 2008).
4.2 Experimental Data
When the same transgenic functional assay used to establish the function of PiSLF 2 in pollen specificity was used to examine two other alleles of PiSLF and several alleles of SLF1 of P. hybrida, the results showed that none of them behaved as expected for the pollen specificity gene. For example, as shown in Fig. 24.1b, introduction of the S 3 -allele of PiSLF, PiSLF 3 , into P. inflata plants of S 2 S 3 genotype did not cause breakdown of SI in S 2 transgenic pollen, even though the transgenic S 2 pollen is heteroallelic (Kubo et al. 2010). Moreover, S 7 -allele of P. hybrida SLF1 caused breakdown of SI in S 9 and S 17 transgenic pollen, but not in S 5 , S 11 , or S 19 transgenic pollen (Kubo et al. 2010). These results, on the one hand, further confirm the involvement of SLF1 in pollen specificity, but on the other hand, suggest that SLF1 is not the only gene involved in pollen specificity. For example, based on the protein degradation model discussed in Sect. 24.3 earlier, the failure of a PiSLF 3 transgene to cause breakdown of SI in S 2 pollen suggests that PiSLF3 does not interact with S2-RNase, a non-self S-RNase, to mediate its ubiquitination and degradation (Fig. 24.1d). This notion raises a question as to whether PiSLF3 interacts with any other non-self S-RNases. Most importantly, this finding suggests that there must be additional protein(s) produced in S 3 pollen that can interact with and detoxify S 2 -RNase, as S 3 pollen is compatible with S 2 -carrying pistils.
The most definitive evidence for the involvement of additional gene(s) in pollen specificity came from the finding that the deduced amino-acid sequences of the S7-allele of SLF1 of P. hybrida and S 9 -allele of SLF1 of P. axillaris are completely identical, even though S 7 and S 9 are genetically distinct S-haplotypes and the deduced amino-acid sequences of their S-RNase alleles are only 45 % identical (Kubo et al. 2010).
5 Involvement of Additional SLF Genes in Controlling Pollen Specificity
After the identification of the first SLF gene, SLF1, it was discovered that there exist additional F-box genes that are tightly linked to the S-locus. For example, our lab identified four such PiSLF-like genes, named PiSLFLa, PiSLFLb, PiSLFLc, and PiSLFLd (Hua et al. 2007). Interestingly, these genes are also specifically expressed in pollen, and at least three of them show allelic-specific sequence differences. However, these PiSLF-like genes were initially thought not to be involved in pollen specificity, as when PiSLFLc-S 1 , PiSLFLb-S 2 , and PiSLFLd-S 2 were introduced into S 1 S 2 , S 2 S 3 , and S 2 S 3 , respectively, none of them caused breakdown of SI in S 2 pollen (in the case of PiSLFLc-S 1 ) or S 3 pollen (in the case of PiSLFLb-S 2 and PiSLFLd-S 2 ) (Hua et al. 2007). The likely involvement of additional gene(s) in pollen specificity prompted the reexamination of these PiSLF-like genes, as well as additional SLF1-like genes of P. hybrida identified by the laboratory of Seiji Takayama.
Takayama’s lab used PCR primers designed based on the sequences of PiSLFLb-S 2 , PiSLFLc-S 1 , and PiSLFLd-S 2 to identify 30 SLF-like genes from different S-haplotypes of P. hybrida, and classified them into five types, type 2 SLF (or SLF2) to type 6 SLF (SLF6) (Kubo et al. 2010). Within each type, the sequence identity between different alleles is high, ranging from 70.3 % to 99 %. However, the sequence identity between different types is only ~50 %. To date, SLF2 and SLF3 have been shown to be involved in pollen specificity as well. For example, S 7 -SLF2 (SLF2 of S 7 haplotype) caused breakdown of SI in S 9 , S 11 , and S 19 pollen, and S 11 -SLF3 (SLF3 of S 11 haplotype) caused breakdown of SI in S 7 pollen. The results also suggest that additional SLF genes are required for pollen specificity. For example, SLF1, SLF2, and SLF3 of S 7 -haplotype did not cause breakdown of SI in S 5 pollen (Kubo et al. 2010).
6 Collaborative Non-Self Recognition Model
After the discovery that multiple SLF proteins are involved in pollen specificity, a modified protein degradation model, named collaborative non-self recognition, was proposed. This model proposes that, for a given S-haplotype, each type of SLF protein interacts with a subset of non-self S-RNases and that multiple types of SLF proteins are required to recognize the entire suite of non-self S-RNases to mediate their degradation. Similar to the original protein degradation model, the new model also predicts that none of the SLF proteins produced in pollen of a given S-haplotype interacts with their self-S-RNase. The prediction that an SLF interacts with a subset of its non-self S-RNases was confirmed by a co-immunoprecipitation (co-IP) experiment. This experiment showed that S7-SLF2:FLAG produced in transgenic pollen coprecipitated, by an anti-FLAG antibody, with S9-RNase and S11-RNase, but not with S5-RNase or S7-RNase, in style extracts. Moreover, the transgenic functional assay showed that expression of S 7 -SLF2 caused breakdown of SI in S 9 and S 11 pollen, but did not cause breakdown of SI in S 5 or S 7 pollen (Kubo et al. 2010). Thus, the co-IP results not only provide strong support for the protein degradation aspect of collaborative non-self recognition, but also are entirely consistent with the results of the transgenic functional assay.
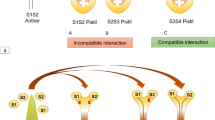
The collaborative non-self recognition model is graphically illustrated in Fig. 24.2. This figure depicts the outcomes of pollination of S 5 -, S 17 -, S 11 -, and S 9 -carrying pistils by S 5 pollen, as explained by the relationships between two types of SLF proteins produced in S 5 pollen, S5-SLF1 and S5-SLF2, and four S-RNases, S5-, S17-, S11-, and S9-RNases, based on the results obtained from the transgenic functional assay (Kubo et al. 2010). When S5-RNase is taken up by the S 5 pollen tube, none of the SLF proteins produced in S 5 pollen can interact with their self-S-RNase, S5-RNase; thus, pollen RNAs are degraded by S5-RNase and pollen tube growth is inhibited. When S17-RNase is taken up by the S 5 pollen tube, S5-SLF1 interacts with S17-RNase to mediate its ubiquitination/degradation. When S11-RNase is taken up by the S 5 pollen tube, S5-SLF2 interacts with S11-RNase to mediate its ubiquitination/degradation. When S9-RNase is taken up by the S 5 pollen tube, both S5-SLF1 and S5-SLF2 will be able to interact with S9-RNase to mediate its ubiquitination/degradation. Thus, in cross-pollinations, at least one SLF protein can interact with and detoxify a non-self S-RNase, allowing the pollen tube to grow down through the style to effect fertilization.
Collaborative non-self recognition model. For pollen of a given S-haplotype, multiple types of SLF proteins collaboratively interact with all their non-self S-RNases, with each type of SLF only interacting with a subset of the non-self S-RNases. For the sake of simplicity, only two different types of SLF proteins, SLF1 and SLF2, are shown for S 5 pollen. When S5-RNase is taken up by the S 5 pollen tube (leftmost panel), no SLF proteins can interact with their self-S-RNase, S5-RNase. Pollen RNA is degraded by S5-RNase and pollen tube growth is inhibited. When S17-RNase is taken up by the S 5 pollen tube (second panel from left), S5-SLF1 interacts with S17-RNase to mediate its degradation. When S11-RNase is taken up by the S 5 pollen tube (second panel from right), S5-SLF2 interacts with S11-RNase to mediate its degradation. When S9-RNase is taken up by the S 5 pollen tube (rightmost panel), both S5-SLF1 and S5-SLF2 interact with S9-RNase to mediate its degradation. Degradation of S17-RNase, S11-RNase, and S9-RNase inside S 5 pollen tubes mediated by one or more types of SLF proteins explains compatible pollinations between S 5 pollen and S 17 -, S 11 -, and S 9 -carrying pistils
The collaborative non-self recognition model can explain the puzzles about the properties of SLF1 mentioned in Sect. 24.4.1, for example, the low allelic sequence diversity of SLF1. As multiple types of SLF proteins act collaboratively to interact with all non-self S-RNases, each SLF only has to interact with a small number of non-self S-RNases, and thus it is unnecessary to have a high degree of allelic sequence diversity. In extreme cases, some allelic variants of an SLF can have identical amino-acid sequences, as is the case for S7-SLF1 and S19-SLF1 of P. axillaris (Kubo et al. 2010). In this case, S 7 and S 19 pollen most likely use their SLF1 proteins (with identical sequences) to interact with the same subset of their common non-self S-RNases (i.e., not including S7-RNase or S19-RNase). S 7 and S 19 pollen could then use some other SLF protein(s) with allelic sequence diversity, or use different types of SLF proteins, to recognize and detoxify S19-RNase and S7-RNase, respectively. Indeed, the degree of amino-acid sequence diversity between different types of SLF proteins is comparable to that of S-RNase, allowing different types of SLF proteins to interact with different S-RNases.
7 Conclusions
Molecular studies of S-RNase-based SI over the preceding more than a quarter century have revealed a number of unexpected findings, suggesting that this inbreeding-prevention genetic system is more complex than initially thought. The most unexpected findings are (1) pollen specificity comprises multiple SLF genes, and (2) rejection of self-pollen is the result of non-self recognition between SLFs and S-RNases, and not by specific self-interaction between an SLF and its cognate S-RNase. Thus, the mechanism of S-RNase-based SI is fundamentally different from the mechanisms of Brassicaceae and Papaveraceae SI, as each employs a single male S-gene and a single female S-gene to control SI, and both involve the interaction between products of matching alleles of the pollen and pistil S-genes (Iwano and Takayama 2012).
The finding that pollen specificity is controlled by multiple SLF proteins has also raised a number of interesting and challenging questions about S-RNase-based SI. We list some of the questions here. For a given S-haplotype, how many SLF genes constitute pollen specificity? Do all S-haplotypes employ the same number of SLF genes? What is the mechanism for the generation of SLF genes? What is the biochemical basis for the differential interactions between SLF proteins and S-RNases? Why does this SI system employ a single gene with a very high degree of allelic sequence polymorphism for pistil specificity, and multiple genes, each with a much lower degree of allelic sequence polymorphism for pollen specificity, but not vice versa? How have a single S-RNase gene and multiple polymorphic SLF genes coevolved to maintain SI?
Efforts are under way to address some of these questions. For example, our laboratory is using RNA-seq to characterize the complete pollen transcriptome of S 2 and S 3 haplotypes of P. inflata to identify all the F-box genes that are linked to the S-locus. The genes identified could then be examined by the well-established transgenic functional assay (Sijacic et al. 2004; Hua et al. 2007; Kubo et al. 2010) to see whether they can cause breakdown of SI in pollen of any non-self S-haplotype. As more SLF genes are identified, comparison of deduced amino-acid sequences of different alleles of the same type of SLF, and deduced amino-acid sequences of different types of SLF genes will likely reveal amino-acid sequences/domains of SLFs that are involved in allele specificity and in type specificity. The approach of chimeric genes could then be used to identify these sequence features. S-RNase-based SI also provides a model for F-box protein-mediated protein degradation, a key regulatory mechanism in many cellular and developmental processes in eukaryotes. Identification of most, if not all, SLF genes of S 2 - and S 3 -haplotype involved in pollen specificity and understanding of how these SLF proteins interact with their specific S-RNases will lay the foundation for studying the structural basis of interactions between F-box proteins and their substrates.
References
Ai YJ, Singh A, Coleman CE, Ioerger TR, Kheyrpour A, Kao T-H (1990) Self-incompatibility in Petunia inflata—isolation and characterization of cDNAs encoding 3 S-allele-associated proteins. Sex Plant Reprod 3(2):130–138
Anderson MA, Cornish EC, Mau SL, Williams EG, Hoggart R, Atkinson A, Bonig I, Grego B, Simpson R, Roche PJ, Haley JD, Penschow JD, Niall HD, Tregear GW, Coghlan JP, Crawford RJ, Clarke AE (1986) Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature (Lond) 321(6065):38–44
Bredemeijer GMM, Blaas J (1981) S-specific proteins in styles of self-incompatible Nicotiana alata. Theor Appl Genet 59(3):185–190. doi:10.1007/bf00264974
Brewbaker JL, Natarajan AT (1960) Centric fragments and pollen-part mutation of incompatibility alleles in Petunia. Genetics 45(6):699–704
Broothaerts W, Vanvinckenroye P, Decock B, Vandamme J, Vendrig JC (1991) Petunia hybrida S-proteins: ribonuclease activity and the role of their glycan side chains in self-incompatibility. Sex Plant Reprod 4(4):258–266
Dowd PE, McCubbin AG, Wang X, Verica JA, Tsukamoto T, Ando T, Kao T-H (2000) Use of Petunia inflata as a model for the study of solanaceous type self-incompatibility. Ann Bot 85:87–93. doi:10.1006/anbo.1999.1032
Entani T, Takayama S, Iwano M, Shiba H, Che FS, Isogai A (1999) Relationship between polyploidy and pollen self-incompatibility phenotype in Petunia hybrida Vilm. Biosci Biotechnol Biochem 63(11):1882–1888
Entani T, Iwano M, Shiba H, Che FS, Isogai A, Takayama S (2003) Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8(3):203–213
Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez-Santana S, Kim S, Phillips TE, Cruz-Garcia F, McClure B (2006) Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature (Lond) 439(7078):805–810. doi:10.1038/Nature04491
Golz JF, Su V, Clarke AE, Newbigin E (1999) A molecular description of mutations affecting the pollen component of the Nicotiana alata S locus. Genetics 152(3):1123–1135
Hua ZH, Kao T-H (2006) Identification and characterization of components of a putative Petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18(10):2531–2553. doi:10.1105/tpc.106.041061
Hua Z, Kao T-H (2008) Identification of major lysine residues of S3-RNase of Petunia inflata involved in ubiquitin-26S proteasome-mediated degradation in vitro. Plant J 54(6):1094–1104. doi:10.1111/j.1365-313X.2008.03487.x
Hua ZH, Meng XY, Kao T-H (2007) Comparison of Petunia inflata S-locus F-box protein (Pi SLF) with Pi SLF-like proteins reveals its unique function in S-RNase-based self-incompatibility. Plant Cell 19(11):3593–3609. doi:10.1105/tpc.107.055426
Hua ZH, Fields A, Kao T-H (2008) Biochemical models for S-RNase-based self-incompatibility. Mol Plant 1(4):575–585. doi:10.1093/Mp/Ssn032
Huang S, Lee HS, Karunanandaa B, Kao T-H (1994) Ribonuclease activity of Petunia inflata S-proteins is essential for rejection of self-pollen. Plant Cell 6(7):1021–1028
Ida K, Norioka S, Yamamoto M, Kumasaka T, Yamashita E, Newbigin E, Clarke AE, Sakiyama F, Sato M (2001) The 1.55 Å resolution structure of Nicotiana alata SF11-RNase associated with gametophytic self-incompatibility. J Mol Biol 314(1):103–112. doi:10.1006/jmbi.2001.5127
Ioerger TR, Clark AG, Kao T-H (1990) Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc Natl Acad Sci USA 87(24):9732–9735. doi:10.1073/pnas.87.24.9732
Ioerger TR, Gohlke JR, Xu B, Kao T-H (1991) Primary structural features of the self-incompatibility protein in Solanaceae. Sex Plant Reprod 4(2):81–87
Iwano M, Takayama S (2012) Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol 15(1):78–83. doi:10.1016/j.pbi.2011.09.003
Kao T-H, McCubbin AG (1996) How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc Natl Acad Sci USA 93(22):12059–12065. doi:10.1073/pnas.93.22.12059
Kao T-H, Tsukamoto T (2004) The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16:S72–S83. doi:10.1105/Tpc.016154
Karunanandaa B, Huang S, Kao T-H (1994) Carbohydrate moiety of the Petunia inflata S3 protein is not required for self-incompatibility interactions between pollen and pistil. Plant Cell 6(12):1933–1940. doi:10.1105/tpc.6.12.1933
Kawata Y, Sakiyama F, Tamaoki H (1988) Amino-acid sequence of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem 176(3):683–697
Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua ZH, Toyoda M, Kawashima S, Ando T, Isogai A, Kao T-H, Takayama S (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330(6005):796–799. doi:10.1126/science.1195243
Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50(1):29–42
Lee HS, Huang S, Kao T-H (1994) S proteins control rejection of incompatible pollen in Petunia inflata. Nature (Lond) 367(6463):560–563. doi:10.1038/367560a0
Liu B, Morse D, Cappadocia M (2009) Compatible pollinations in Solanum chacoense decrease both S-RNase and S-RNase mRNA. PLoS One 4(6):e5774. doi:10.1371/journal.pone.0005774
Luu DT, Qin XK, Morse D, Cappadocia M (2000) S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature (Lond) 407(6804):649–651
Matsuura T, Sakai H, Unno M, Ida K, Sato M, Sakiyama F, Norioka S (2001) Crystal structure at 1.5-Å resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility. J Biol Chem 276(48):45261–45269. doi:10.1074/jbc.M107617200
Matton DP, Maes O, Laublin G, Xike Q, Bertrand C, Morse D, Cappadocia M (1997) Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9(10):1757–1766. doi:10.1105/tpc.9.10.1757
Matton DP, Luu DT, Xike Q, Laublin G, O’Brien M, Maes O, Morse D, Cappadocia M (1999) Production of an S-RNase with dual specificity suggests a novel hypothesis for the generation of new S alleles. Plant Cell 11(11):2087–2097
McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE (1989) Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature (Lond) 342(6252):955–957
McCubbin AG, Kao T-H (2000) Molecular recognition and response in pollen and pistil interactions. Annu Rev Cell Dev Biol 16:333–364. doi:10.1146/annurev.cellbio.16.1.333
McCubbin AG, Wang X, Kao T-H (2000) Identification of self-incompatibility (S-) locus linked pollen cDNA markers in Petunia inflata. Genome 43(4):619–627. doi:10.1139/Gen-43-4-619
Murfett J, Atherton TL, Mou B, Gasser CS, McClure BA (1994) S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature (Lond) 367(6463):563–566. doi:10.1038/367563a0
Newbigin E, Paape T, Kohn JR (2008) RNase-based self-incompatibility: puzzled by Pollen S. Plant Cell 20(9):2286–2292. doi:10.1105/tpc.108.060327
Paape T, Igic B, Smith SD, Olmstead R, Bohs L, Kohn JR (2008) A 15-Myr-old genetic bottleneck. Mol Biol Evol 25(4):655–663. doi:10.1093/molbev/msn016
Sato K, Nishio T, Kimura R, Kusaba M, Suzuki T, Hatakeyama K, Ockendon DJ, Satta Y (2002) Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics 162(2):931–940
Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao T-H (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature (Lond) 429(6989):302–305. doi:10.1038/Nature02523
Singh A, Ai Y, Kao T-H (1991) Characterization of ribonuclease activity of three S-allele-associated proteins of Petunia inflata. Plant Physiol 96(1):61–68
Stout AB, Chandler C (1942) Hereditary transmission of induced tetraploidy and compatibility in fertilization. Science 96:257–258. doi:10.1126/science.96.2489.257-a
Takayama S, Isogai A (2005) Self-incompatibility in plants. Annu Rev Plant Biol 56:467–489. doi:10.1146/annurev.arplant.56.032604.144249
Takebayashi N, Brewer PB, Newbigin E, Uyenoyama MK (2003) Patterns of variation within self-incompatibility loci. Mol Biol Evol 20(11):1778–1794. doi:10.1093/molbev/msg209
Tsai DS, Lee HS, Post LC, Kreiling KM, Kao T-H (1992) Sequence of an S-protein of Lycopersicon peruvianum and comparison with other solanaceous S-proteins. Sex Plant Reprod 5(4):256–263
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15(3):771–781
Wang H-Y, Xue Y-B (2005) Subcellular localization of the S locus F-box protein AhSLF-S2 in pollen and pollen tubes of self-incompatible Antirrhinum. J Integr Plant Biol 47(1):76–83. doi:10.1111/j.1744-7909.2005.00014.x
Wang X, Hughes AL, Tsukamoto T, Ando T, Kao T-H (2001) Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol 125(2):1012–1022. doi:10.1104/pp. 125.2.1012
Wang Y, Wang X, McCubbin AG, Kao T-H (2003) Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflata. Plant Mol Biol 53(4):565–580. doi:10.1023/B:Plan.0000019068.00034.09
Wang Y, Tsukamoto T, Yi KW, Wang X, Huang SS, McCubbin AG, T-H K (2004) Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881-kb contig containing S 2 -RNase. Plant Mol Biol 54(5):727–742. doi:10.1023/B:Plan.0000040901.98982.82
Zhou JL, Wang F, Ma WS, Zhang YS, Han B, Xue YB (2003) Structural and transcriptional analysis of S-locus F-box genes in Antirrhinum. Sex Plant Reprod 16(4):165–177. doi:10.1007/s00497-003-0185-5
Zurek DM, Mou BQ, Beecher B, McClure B (1997) Exchanging sequence domains between S-RNases from Nicotiana alata disrupts pollen recognition. Plant J 11(4):797–808. doi:10.1046/j.1365-313X.1997.11040797.x
Acknowledgments
The work from the authors’ laboratory was supported by grants to T.-H. K. from the U.S. National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
This chapter is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
Copyright information
© 2014 The Author(s)
About this paper
Cite this paper
Sun, P., Williams, J.S., Li, S., Kao, Th. (2014). S-RNase-Based Self-Incompatibility in Petunia: A Complex Non-Self Recognition System Between Pollen and Pistil. In: Sawada, H., Inoue, N., Iwano, M. (eds) Sexual Reproduction in Animals and Plants. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54589-7_24
Download citation
DOI: https://doi.org/10.1007/978-4-431-54589-7_24
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54588-0
Online ISBN: 978-4-431-54589-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)