Abstract
Key message
Arabidopsis det1 mutants exhibit salt and osmotic stress resistant germination. This phenotype requires HY5, ABF1, ABF3, and ABF4.
Abstract
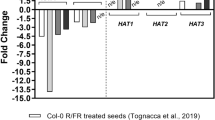
While DE-ETIOLATED 1 (DET1) is well known as a negative regulator of light development, here we describe how det1 mutants also exhibit altered responses to salt and osmotic stress, specifically salt and mannitol resistant germination. LONG HYPOCOTYL 5 (HY5) positively regulates both light and abscisic acid (ABA) signalling. We found that hy5 suppressed the det1 salt and mannitol resistant germination phenotype, thus, det1 stress resistant germination requires HY5. We then queried publically available microarray datasets to identify genes downstream of HY5 that were differentially expressed in det1 mutants. Our analysis revealed that ABA regulated genes, including ABA RESPONSIVE ELEMENT BINDING FACTOR 3 (ABF3), are downregulated in det1 seedlings. We found that ABF3 is induced by salt in wildtype seeds, while homologues ABF4 and ABF1 are repressed, and all three genes are underexpressed in det1 seeds. We then investigated the role of ABF3, ABF4, and ABF1 in det1 phenotypes. Double mutant analysis showed that abf3, abf4, and abf1 all suppress the det1 salt/osmotic stress resistant germination phenotype. In addition, abf1 suppressed det1 rapid water loss and open stomata phenotypes. Thus interactions between ABF genes contribute to det1 salt/osmotic stress response phenotypes.










Similar content being viewed by others
Abbreviations
- ABF:
-
Abscisic acid responsive element binding factor
- ABI5:
-
Abscisic acid insensitive 5
- DET1:
-
De-etiolated 1
- HY5:
-
Long hypocotyl 5
References
Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110:E435–E444
Bhattacharjee S, Saha AK (2014) Plant water-stress response mechanisms. In: Approaches to plant stress and their management. Springer, pp 149–172
Bolle C (2009) Phenotyping of abiotic responses and hormone treatments in Arabidopsis. Plant Signal Transduct 479:35–59
Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318:801–806
Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37:425–435
Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10:673–683
Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, Xiong L (2008) Integration of light and abscisic acidsignalling during seed germination and early seedling development. Proc Natl Acad Sci USA 105:4495–4500
Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim B, Lee S, Kudla J, Luan S (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52:223–239
Choi HI, Hong JH, Ha JO, Kang JY, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275:1723–1730
Chory J (1992) A genetic model for light-regulated seedling development in Arabidopsis. Development 115:337–354
Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19:371–379
Delgado D, Ballesteros I, Torres-Contreras J, Mena M, Fenoll C (2012) Dynamic analysis of epidermal cell divisions identifies specific roles for COP10 in Arabidopsis stomatal lineage development. Planta 236:447–461
Fernando VCD, Schroeder DF (2015) Genetic interactions between DET1 and intermediate genes in Arabidopsis ABA signalling. Plant Sci 239:166–179
Fernando VCD, Schroeder DF (2016) Arabidopsis DDB1-CUL4 E3 ligase complexes in det1 salt/osmotic stress resistant germination. Plant Signal Behav 11:e1223004
Finkelstein R, Gampala SS, Lynch TJ, Thomas TL, Rock CD (2005) Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE (ABI) 5 and ABRE-BINDING FACTOR (ABF) 3. Plant Mol Biol 59:253–267
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17:3470–3488
Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V (2008) CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res 36:D986–D990
Hossain Z, Amyot L, McGarvey B, Gruber M, Jung J, Hannoufa A (2012) The translation elongation factor eEF-1Bβ1 is involved in cell wall biosynthesis and plant development in Arabidopsis thaliana. PLoS ONE 7:e30425
Huang X, Ouyang X, Deng XW (2014) Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signalling. Curr Opin Plant Biol 21:96–103
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14:343–357
Kang C, Lian H, Wang F, Huang J, Yang HQ, Kang C, Lian HL, Wang F, Huang J, Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21:2624–2641
Khanna R, Li J, Tseng T, Schroeder JI, Ehrhardt DW, Briggs WR (2014) COP1 jointly modulates cytoskeletal processes and electrophysiological responses required for stomatal closure. Mol Plant 7:1441–1454
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363
Kim S, Kang J, Cho D, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signalling and its overexpression affects multiple stress tolerance. Plant J 40:75–87
Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19:731–749
Li X, Ma XG, He JM (2013) Stomatal bioassay in Arabidopsis leaves. Bio-protocol 3:921
Mackinney G (1941) Absorption of light by chlorophyll solutions. J Biol Chem 140:315–322
Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102:12270–12275
Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126:467–475
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466
Pepper AE, Chory J (1997) Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics 145:1125–1137
Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15:395–401
Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J (2006) TM4 microarray software suite. Meth Enzymol 411:134–193
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Tholakalabavi A, Zwiazek JJ, Thorpe TA (1994) Effect of mannitol and glucose-induced osmotic stress on growth, water relations, and solute composition of cell suspension cultures of poplar (Populus deltoides var. Occidentalis) in relation to anthocyanin accumulation. In Vitro Cell Dev Biol Plant 30:164–170
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97:11632–11637
Weigel D, Glazebrook J (2002) Arabidopsis: a laboratory manual. CSHL Press, New York
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signalling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685
Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ 38:35–49
Yu Y, Wang J, Shi H, Gu J, Dong J, Deng XW, Huang R (2016) Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol 170:2340–2350
Acknowledgements
This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Author information
Authors and Affiliations
Contributions
WAK performed preliminary experiments, MFB analyzed microarray data. All other experiments were performed by VCDF. All authors contributed to experimental design, manuscript writing, and editing.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fernando, V.C.D., Al Khateeb, W., Belmonte, M.F. et al. Role of Arabidopsis ABF1/3/4 during det1 germination in salt and osmotic stress conditions. Plant Mol Biol 97, 149–163 (2018). https://doi.org/10.1007/s11103-018-0729-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0729-6