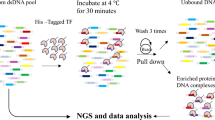
Abstract
The interaction between a protein and DNA is involved in almost all cellular functions, and is vitally important in cellular processes. Two complementary approaches are used to detect the interactions between a transcription factor (TF) and DNA, i.e. the TF-centered or protein–DNA approach, and the gene-centered or DNA–protein approach. The yeast one-hybrid (Y1H) is a powerful and widely used system to identify DNA–protein interactions. However, a powerful method to study protein–DNA interactions like Y1H is lacking. Here, we developed a protein–DNA method based on the Y1H system to identify the motifs recognized by a defined TF, termed TF-centered Y1H. In this system, a random short DNA sequence insertion library was generated as the prey DNA sequences to interact with a defined TF as the bait. Using this system, novel interactions were detected between DNA motifs and the AtbZIP53 protein from Arabidopsis. We identified six motifs that were specifically bound by AtbZIP53, including five known motifs (DOF, G-box, I-box, BS1 and MY3) and a novel motif BRS1 [basic leucine zipper (bZIP) Recognized Site 1]. The different subfamily bZIP members also recognize these six motifs, further confirming the reliability of the TF-centered Y1H results. Taken together, these results demonstrated that TF-centered Y1H could identify quickly the motifs bound by a defined TF, representing a reliable and efficient approach with the advantages of Y1H. Therefore, this TF-centered Y1H may have a wide application in protein–DNA interaction studies.







Similar content being viewed by others
References
Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21:1747–1761
Alves MS, Dadalto SP, Gonçalves AB, De Souza GB, Barros VA, Fietto LG (2013) Plant bZIP transcription factors responsive to pathogens: a review. Int J Mol Sci 14:7815–7828
Arda HE, Walhout AJ (2010) Gene-centered regulatory networks. Brief Funct Genomics 9:4–12
Dey B, Thukral S, Krishnan S, Chakrobarty M, Gupta S, Manghani C, Rani V (2012) DNA–protein interactions: methods for detection and analysis. Mol Cell Biochem 365:279–299
Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W (2011) Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23:381–395
Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3:11
Hwang YS, Karrer EE, Thomas BR, Chen L, Rodriguez RL (1998) Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Mol Biol 36:331–341
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: betaglucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Kim TH, Ren B (2007) Genome-wide analysis of protein–DNA interactions. Annu Rev Genomics Hum Genet 7:81–102
Lacombe E, Van Doorsselaere J, Boerjan W, Boudet AM, Grima-Pettenati J (2000) Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA reductase gene and for protein–DNA complex formation. Plant J 23:663–676
Lee SC, Choi HW, Hwang IS, du Choi S, Hwang BK (2006) Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 224:1209–1225
Mallappa C, Yadav V, Negi P, Chattopadhyay S (2006) A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J Biol Chem 281:22190–22199
Meng X, Brodsky MH, Wolfe SA (2005) A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat Biotechnol 23:988–994
Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, Shinozaki MFK, Matsui M, Ohme-Takagi M (2010) Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol 51:2145–2151
Nadja B, Claus F, Salome P, Lothar W (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J 13:5383–5392
Oliphant AR, Brandl CJ, Struhl K (1989) Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 proteins. Mol Cell Biol 9:2944–2949
Reece-Hoyes JS, Barutcu AR, McCord RP, Jeong JS, Jiang L, MacWilliams A, Yang X, Salehi-Ashtiani K, Hill DE, Blackshaw S, Zhu H, Dekker J, Walhout AJ (2011) Yeast one-hybrid assays for gene-centered human gene regulatory network mapping. Nat Methods 8:1050–1052
Rose A, Meier I, Wienand U (1999) The tomato I-box binding factor LeMYBI is a member of a novel class of myb-like proteins. Plant J 20:641–652
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Van SB, Henikoff S (2000) Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol 18:424–428
Vinson CR, Sigler PB, McKnight SL (1989) Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 246:911–916
Vogel MJ, Peric-Hupkes D, Van SB (2007) Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nat Protoc 2:1467–1478
Walhout AJM (2006) Unraveling transcription regulatory networks by protein–DNA and protein–protein interaction mapping. Genome Res 16:1445–1454
Wei Z, Angerer RC, Angerer LM (1999) Identification of a new sea urchin ets protein, SpEts4, by yeast one-hybrid screening with the hatching enzyme promoter. Mol Cell Biol 19:1271–1278
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683
Acknowledgments
This work was supported by The Hundred Talents Program of the Chinese Academy of Sciences. We are grateful to our colleagues who have shared their knowledge and reagents, the State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiaoyu Ji and Liuqiang Wang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ji, X., Wang, L., Nie, X. et al. A novel method to identify the DNA motifs recognized by a defined transcription factor. Plant Mol Biol 86, 367–380 (2014). https://doi.org/10.1007/s11103-014-0234-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0234-5