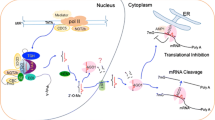
Abstract
MicroRNAs (miRNAs) are ~21-nucleotide long endogenous small RNAs that regulate gene expression through post-transcriptional or transcriptional gene silencing and/or translational inhibition. miRNAs can arise from the “exon” of a MIRNA gene, from an intron (e.g. mirtrons in animals), or from the antisense strand of a protein coding gene (natural antisense microRNAs, nat-miRNAs). Here we demonstrate that two functionally related miRNAs, miR842 and miR846, arise from the same transcription unit but from alternate splicing isoforms. miR846 is expressed only from Isoform1 while in Isoforms2 and -3, a part of pre-miR846 containing the miRNA* sequence is included in the intron. The splicing of the intron truncates the pre-MIRNA and disrupts the expression of the mature miR846. We name this novel phenomenon splicing-regulated miRNA. Abscisic acid (ABA) is shown to mediate the alternative splicing event by reducing the functional Isoform1 and increasing the non-functional Isoform3, thus repressing the expression of miR846 concomitant with accumulation of an ABA-inducible target jacalin At5g28520 mRNA, whose cleavage was shown by modified 5′-RACE. This regulation shows the functional importance of splicing-regulated miRNA and suggests possible mechanisms for altered ABA response phenotypes of miRNA biogenesis mutants. Arabidopsis lyrata-MIR842 and Aly-MIR846 have conserved genomic arrangements with A. thaliana and candidate target jacalins, similar primary transcript structures and intron processing, and better miRNA–miRNA* pairings, suggesting that the interactions between ABA, MIR842, MIR846 and jacalins are similar in A. lyrata. Together, splicing-regulated miRNAs, nat-miRNAs/inc-miRNAs and mirtrons illustrate the complexity of MIRNA genes, the importance of introns in the biogenesis and regulation of miRNAs, and raise questions about the processes and molecular mechanisms that drive MIRNA evolution.





Similar content being viewed by others
References
Allen E, Xie ZX, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36:1282–1290
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H (2005) Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33:2697–2706
Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127:565–577
Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC (2008) Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci U S A 105:7964–7969
Bailey CD, Koch MA, Mayer M, Mummenhoff K, O’Kane SL, Warwick SI, Windham MD, Al-Shehbaz IA (2006) Toward a global phylogeny of the Brassicaceae. Mol Biol Evol 23:2142–2160
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J 17:170–180
Breakfield NW, Corcoran DL, Petricka JJ, Shen J, Sae-Seaw J, Rubio-Somoza I, Weigel D, Ohler U, Benfey PN (2012) High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res 22:163–176
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190
Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q et al (2007) Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131:1340–1353
Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H (2010) siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 38:6883–6894
Chen X, Liu J, Cheng Y, Jia D (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129:1085–1094
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W159
Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16:939–944
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL et al (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219
Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D et al (2010) MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22:1074–1089
Fernandez-Valverde SL, Taft RJ, Mattick JS (2010) Dynamic isomiR regulation in Drosophila development. RNA 16:1881–1888
Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC (2010) MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell 38:900–907
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432:235–240
Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14:854–866
Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17:1376–1386
Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21:3119–3132
Hugouvieux V, Kwak JM, Schroeder JI (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106:477–487
Inan G, Zhang Q, Li PH, Wang ZL, Cao ZY, Zhang H, Zhang CQ, Quist TM, Goodwin SM, Zhu JH et al (2004) Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135:1718–1737
Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126:5231–5243
Jeong DH, Green PJ (2012) Methods for validation of miRNA sequence variants and the cleavage of their targets. Methods. doi:10.1016/j.ymeth.2012.08.005
Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23:4185–4207
Jia X, Wang WX, Ren L, Chen QJ, Mendu V, Willcut B, Dinkins R, Tang X, Tang G (2009) Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol Biol 71:51–59
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kim YK, Kim VN (2007) Processing of intronic microRNAs. EMBO J 26:775–783
Kim VN, Nam JW (2006) Genomics of microRNA. Trends Genet 22:165–173
Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH (2008) Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol 49:1634–1644
Kinoshita N, Wang H, Kasahara H, Liu J, Macpherson C, Machida Y, Kamiya Y, Hannah MA, Chua NH (2012) IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell. doi: 10.1105/tpc.112.097006
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157
Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci U S A 101:12753–12758
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105:8795–8800
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S et al (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060
Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K (2010) Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 16:2170–2180
Li W, Cui X, Meng Z, Huang X, Xie Q, Wu H, Jin H, Zhang D, Liang W (2012) Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol 158:1279–1292
Llave C, Xie ZX, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J (2006) SERRATE: a new player on the plant microRNA scene. EMBO Rep 7:1052–1058
Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12:2351–2366
Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D et al (2008) Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc Natl Acad Sci U S A 105:4951–4956
Luo QJ, Samanta MP, Koksal F, Janda J, Galbraith DW, Richardson CR, Ou-Yang F, Rock CD (2009) Evidence for antisense transcription associated with microRNA target mRNAs in Arabidopsis. PLoS Genet 5:e1000457
Ma Z, Coruh C, Axtell MJ (2010) Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22:1090–1103
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Meng Y, Shao C (2012) Large-scale identification of mirtrons in Arabidopsis and rice. PLoS ONE 7:e31163
Meng Y, Ma X, Chen D, Wu P, Chen M (2010) MicroRNA-mediated signaling involved in plant root development. Biochem Biophys Res Commun 393:345–349
Merchan F, Boualem A, Crespi M, Frugier F (2009) Plant polycistronic precursors containing non-homologous microRNAs target transcripts encoding functionally related proteins. Genome Biol 10:R136
Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C et al (2008) Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127
Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M et al (2008) Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18:610–621
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Neilsen CT, Goodall GJ, Bracken CP (2012) IsomiRs—the overlooked repertoire in the dynamic microRNAome. Trends Genet. doi:10.1016/j.tig.2012.07.005
Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130:89–100
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B (2012) Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci U S A 109:12817–12821
Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49:592–606
Rock CD, Sun X (2005) Crosstalk between ABA and auxin signaling pathways in roots of Arabidopsis thaliana (L.) Heynh. Planta 222:98–106
Ruby JG, Jan CH, Bartel DP (2007a) Intronic microRNA precursors that bypass Drosha processing. Nature 448:83–86
Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC (2007b) Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17:1850–1864
Song L, Han MH, Lesicka J, Fedoroff N (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A 104:5437–5442
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Szarzynska B, Sobkowiak L, Pant BD, Balazadeh S, Scheible WR, Mueller-Roeber B, Jarmolowski A, Szweykowska-Kulinska Z (2009) Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res 37:3083–3093
Válóczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z (2004) Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32:e175
Vartanian N, Marcotte L, Giraudat J (1994) Drought rhizogenesis in Arabidopsis thaliana: differential responses of hormonal mutants. Plant Physiol 104:761–767
Vazquez F, Gasciolli V, Crete P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14:346–351
Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY (2005) Control of root cap formation by microRNA-targeted Auxin response factors in Arabidopsis. Plant Cell 17:2204–2216
Wang H, Zhang X, Liu J, Kiba T, Woo J, Ojo T, Hafner M, Tuschl T, Chua NH, Wang XJ (2011) Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J 67:292–304
Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S et al (2008) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 36:D13–D21
Williams L, Carles CC, Osmont KS, Fletcher JC (2005) A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci U S A 102:9703–9708
Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138:2145–2154
Yan K, Liu P, Wu CA, Yang GD, Xu R, Guo QH, Huang JG, Zheng CC (2012) Stress-nduced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell 484:521–531
Yoon EK, Yang JH, Lim J, Kim SH, Kim SK, Lee WS (2010) Auxin regulation of the microRNA390-dependent trans-acting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res 38:1382–1391
Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC et al (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A 105:10073–10078
Zhan X, Wang B, Li H, Liu R, Kalia RK, Zhu JK, Chinnusamy V (2012) Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci U S A 109:18198–18203
Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646
Zhang JF, Yuan LJ, Shao Y, Du W, Yan DW, Lu YT (2008) The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ 31:562–574
Zhang H, Maniar JM, Fire AZ (2011) ‘Inc-miRs’: functional intron-interrupted miRNA genes. Genes Dev 25:1589–1594
Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C (2008) A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res 18:1456–1465
Acknowledgments
The authors thank Qingjun Luo, Amandeep Mittal and Yingwen Jiang for helpful discussions, and the TTU High Performance Computing Center for use of the Hrothgar cluster. This work was supported by the National Institutes of Health grant GM077245 to C.D.R, and a Texas Tech University AT&T Chancellor’s Fellowship to F.J. The funders had no role in the study design, in the collection, analysis, or interpretation of data, or in the writing of the manuscript or decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2013_15_MOESM1_ESM.pptx
GUS staining of wild type (A, C, E) and Pro-MIR842_MIR846:GUS transgenic plants (B, D, F) showing similar background staining in inflorescence (A, B), the junction between siliques and petioles (C, D), and in the midrib of rosette leaves (E, F). (PPTX 28184 kb)
11103_2013_15_MOESM2_ESM.pptx
Arabidopsis virtual expression analysis from microarray transcriptome eFP data (http://bar.utoronto.ca/) for AT5G28520 visualizing root-specific induction for seedlings treated with 300 mM Mannitol, with 150 mM NaCl (A), or treated with ABA in (B). (PPTX 922 kb)
11103_2013_15_MOESM3_ESM.pptx
Alignment between A. thaliana and A. lyrata genomic sequences containing MIR842 and MIR846. Gray lines represent pre-miRNAs. Black boxes represent mature miRNA or miRNA* sequences. Black arrowheads represent GT..AG intron 5’ and 3’ splice boundaries. Alignment performed with tcoffee (www.tcoffee.org). (PPTX 1310 kb)
11103_2013_15_MOESM4_ESM.pptx
A.lyrata jacalin homologs carrying miR846 complementary sites. Numbers above upper sequences represent nt positions in the mRNA, according to Genbank ID annotations. Vertical lines represent perfect matches. “o” represents a G:U wobble. X represents a mismatch. (PPTX 5758 kb)
11103_2013_15_MOESM5_ESM.pptx
Models of (A) a splicing regulated miRNA, (B) a Nat-miRNA/Inc-miRNA, and (C) a mirtron. Thick black lines represent exons; thick gray lines represent introns; thin gray lines represent flanking sequences; stem-loops represent pre-miRNAs; horizontal lines, Watson–Crick base pairs. (PPTX 907 kb)
11103_2013_15_MOESM6_ESM.pptx
A miR846 variant can target AT5G28520. (A) A part of miR846 stem-loop sequence. The mature miR846 sequence is marked in pink. (B) Base pairing between the potential miR846 variant and AT5G28520 showing extended complementarity at the 5′ end. The mature miR846 sequence is marked in pink. Red underline indicates the variant. Numbers represent positions in the mRNA. Vertical lines represent perfect matches. “o” represents a G:U wobble. X represents a mismatch. The thick black line represents the cleavage site shown by modified 5′-RACE. (PPTX 204 kb)
11103_2013_15_MOESM7_ESM.xlsx
Potential targets of miR842 or miR846 identified with “psRNATarget” (http://plantgrn.noble.org/psRNATarget/) (Dai and Zhao, 2011). (XLSX 20 kb)
Rights and permissions
About this article
Cite this article
Jia, F., Rock, C.D. MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol Biol 81, 447–460 (2013). https://doi.org/10.1007/s11103-013-0015-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0015-6