Abstract
Plant ammonium transporters of the AMT1 family are involved in N-uptake from the soil and ammonium transport, and recycling within the plant. Although AMT1 genes are known to be expressed in nitrogen-fixing nodules of legumes, their precise roles in this specialized organ remain unknown. We have taken a reverse-genetic approach to decipher the physiological role of LjAMT1;1 in Lotus japonicus nodules. LjAMT1;1 is normally expressed in both the infected zone and the vascular tissue of Lotus nodules. Inhibition of LjAMT1;1 gene expression, using an antisense gene construct driven by a leghemoglobin promoter resulted in a substantial reduction of LjAMT1;1 transcript in the infected tissue but not the vascular bundles of transgenic plants. As a result, the nitrogen-fixing activity of nodules was partially impaired and nodule number increased compared to control plants. Expression of LjAMT1;1-GFP fusion protein in plant cells indicated a plasma-membrane location for the LjAMT1;1 protein. Taken together, the results are consistent with a role of LjAMT1;1 in retaining ammonium derived from symbiotic nitrogen fixation in plant cells prior to its assimilation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ammonium is a primary source of nitrogen for plants. It is imported into plant cells from the surrounding environment via transporters, in the plasma membrane (PM) of root cells (Yuan et al. 2007b) and leaf cells (Husted and Schjoerring 1996). A large fraction of ammonium assimilated by plants is generated within cells by reduction of nitrate and nitrite obtained from the soil, via the photorespiratory nitrogen cycle in leaf mitochondria and by catabolism of endogenous amino compounds. Ammonium transport across plant membranes is mediated by proteins of the high-affinity ammonium transporter (AMT) (Howitt and Udvardi 2000; von Wiren and Merrick 2004), which are cation (NH4 +) uniporters (Ludewig et al. 2002; Wood et al. 2006). Members of the AMT1 and AMT2 subfamilies differ in their biochemical and sequence characteristics with the latter being more closely related to bacterial AMT transporters (Sohlenkamp et al. 2000). Genome sequencing has revealed three AMT1 genes in Oriza sativa (Sonoda et al. 2003), five in Arabidopsis thaliana and eight in Populus trichocarpa (Couturier et al. 2007); whereas one, six and seven AMT2 genes were identified in A. thaliana (Sohlenkamp et al. 2000) P. trichocarpa (Couturier et al. 2007) and O. sativa (Suenaga et al. 2003), respectively.
Complementary, partially overlapping gene expression patterns and kinetic properties of Arabidopsis AMT1 transporters indicate that they work cooperatively to ensure a coordinate effective high-affinity ammonium uptake in Arabidopsis roots (Yuan et al. 2007b).
Legumes are quite unique amongst plant families in that they can generate ammonium de novo from nitrogen gas (N2), via a symbiosis with nitrogen-fixing bacteria called rhizobia. Symbiotic nitrogen fixation in legumes takes place in specialized organs called root nodules, which develop as a result of a series of signal exchanges between plant root cells and rhizobia in the soil, but only when mineral nitrogen in the soil is limiting for plant growth. Rhizobia take up residence in enlarged cortical cells of nodules, surrounded by a plant membrane known as the symbiosome membrane. Ammonium produced by rhizobia is transported to the host plant cell cytoplasm, via the bacterial inner and outer membranes and the symbiosome membrane. While transport of ammonium across the bacterial membranes may occur via simple diffusion of NH3 through the lipid bilayer (Howitt et al. 1986), distinct membrane proteins are believed to be involved in both NH3 and NH4 + transport across the symbiosome membrane (Udvardi and Day 1997; Niemietz and Tyerman 2000). The molecular identity of the symbiosome membrane NH4 + channel remains unknown. However, the well-known nodulin and aquaporin, Nodulin 26 in soybean and its homologues in other species may provide a channel for NH3 diffusion across the SM (Howitt and Udvardi 2000; Roberts and Tyerman 2002).
AMT family members have been identified in legumes (Salvemini et al. 2001; Simon-Rosin et al. 2003; D’Apuzzo et al. 2004) and characterized at the biochemical, molecular and cellular levels (Simon-Rosin et al. 2003; D’Apuzzo et al. 2004).
LjAMT1;1 and a related gene of the AMT2 sub-family, LjAMT2;1 are expressed in nodules of Lotus japonicus within the infected zone and vascular bundles (Simon-Rosin et al. 2003; D’Apuzzo et al. 2004). However, the physiological role of neither of these has been elucidated. Here, we present results of reverse-genetic experiments that shed light on the role and importance of LjAMT1;1 in Lotus nodules.
Material and methods
Plant material, growth conditions and in vitro nodulation assay
All experiments were carried out with L. japonicus ecotype B-129 F9 GIFU. Growth conditions and Mesorhizobium loti inoculation procedure for the in vitro nodulation assay have been reported elsewhere (Barbulova and Chiurazzi 2005). Seeds were sterilized for 20 min in 25% commercial bleach (1% hypochlorite) and 0.1% Triton, washed six times in sterile H2O and kept over night in water at 4°C. Seeds were sown on semicircles of sterilized filter paper placed on the surface of the 0.1% solidified Jensen medium (Jensen 1942; six seeds per 140 × 10 mm Petri dish) and left over night at 4°C cap side down. After 24 h in the dark in the growth chamber, Petri dishes were exposed to light and kept in a vertical position. Care was taken to maintain the young emerging roots in contact with the filter paper. Unsynchronized seedlings were discarded at this stage. M. loti strain R7A was grown to mid-log phase in liquid TY medium (Beringer 1974) plus 6 mM CaCl2 and used to inoculate plants at a concentration of 107 cells per root tip (root length of about 1.5 cm). Four days after infection, the filter paper was removed and the plants left on the Petri dishes. Aluminum foil was used to keep the roots in the dark. Plants were cultivated in a growth chamber with a light intensity of 200 μmol m−2 s−1 at 23°C with a 16/8 h day/night cycle.
M. loti nifH − was kindly provided by Clive Ronson (University of Otago, New Zealand).
Plant transformation
We followed the procedures described in Handberg and Stougaard (1992) and in Martirani et al. (1999) for Agrobacterium tumefaciens and Agrobacterium rhizogenes mediated plant transformations, respectively.
T-DNA constructs preparation
The 35S-AMT1;1-GFP fusion was prepared in the following way: the LjAMT1;1 cDNA sequence was amplified with the two oligonucleotides 5′-CGCGGATCCATGGCGGCGCTGCCGGAGTG-3′ (including a BamHI site) and 5′-GCGGTACCTGACTCAGCACTAGGAGTGGA-3′ (including a KpnI site), double-digested with BamHI-KpnI and cloned into the β-GFP plasmid (Duby et al. 2001) pre-digested with BamHI-KpnI. The correct sequence for LjAMT1;1-GFP translational fusion was verified by sequencing. The LjAMT1;1-GFP cassette was then cloned as a BamHI-SacI fragment into the pCAMBIA1300 vector pre-digested with BglII-SacI.
The LBC3-AMT1;1 antisense construct was prepared in the following way:
The LjAMT1;1 coding sequence was amplified with two oligonucleotides including EcoRI or BamHI sites, double digested with EcoRI-BamHI and cloned into the pGPTVkan-LBC3 plasmid pre-digested EcoRI-BamHI. The pGPTVkan-LBC3, kindly provided by Mette Groenlund (University of Aahrus, Denmark), is a derivative of the pGPTVkan plasmid (Becker et al. 1992), containing the 1.9 kb fragment of the Glycine max LBC3 promoter cloned into SalI-BamHI sites.
Quantitative real-time RT-PCR
Total RNA was prepared from Lotus tissues using the procedure of Kistner and Matamoros (2005). The samples were treated with DNAse I (Ambion) to remove contaminating DNA the absence of which was subsequently confirmed by PCR. One microgram of total RNA was annealed to random decamers and reverse-transcribed with reverse transcriptase (Ambion) to obtain cDNA. Real time PCR was performed with a DNA Engine Opticon 2 System, MJ Research (MA, USA) using SYBR to monitor dsDNA synthesis. The ubiquitin (UBI) gene (AW719589) was used as an internal standard. The concentration of primers was optimized for each PCR reaction and each amplification was carried out in triplicate. The PCR program used was as follows: 95°C for 13 min and 39 cycles of 94°C for 15 s, 63°C for 15 s and 72°C for 15 s. Data were analyzed using Opticon Monitor Analysis Software Version 2.01 (MJ Research). The relative level of expression was calculated with the following formula: relative expression ratio of the gene of interest is 2−ΔCT with ΔCT = Ctgene minus CTUBI. Analysis of the melting curve of PCR products at the end of the PCR run revealed a single narrow peak for each amplification product, and fragments amplified from total cDNA were gel-purified and sequenced to assure accuracy and specificity.
LjAMT1;1 specific primers were as described by D’Apuzzo et al. (2004).
Histochemical GUS analysis
Histochemical staining of whole plant material was performed as described by Jefferson (1987). After staining, whole roots were fixed with 4% paraformaldehyde, 0.25% glutaraldehyde in 50 mM KPO4 buffer, 5 mM EGTA, 10 mM DTT, pH 7.2 and stored at 4°C. The tissues were then washed with 50 mM KPO4 buffer pH 7.2, embedded in 4% agar and cut into 60 μm sections with a vibratome (Leica VT1000S). Sections were finally analyzed with a light microscope using dark- and bright-field optics.
In situ hybridization
In situ hybridization experiments were performed as previously described (Flemetakis et al. 2004). L. japonicus nodules harvested at 3 weeks post-inoculation with M. loti R7A were fixed in 4% (w/v) paraformaldehyde supplemented with 0.25% (v/v) glutaraldehyde in 10 mM sodium phosphate buffer (pH 7.4) for 4 h in a vacuum aspirator. Fixed nodules were block-stained in 0.5% (w/v) safranin, dehydrated through ethanol series, embedded in paraffin and cut into 8 μm-thin sections. Antisense RNA probe labelled with digoxigenin-11-rUTP (Boehringer Mannheim, Mannheim, Germany) was transcribed from the 300 bp region at the 3′ end of the AMT1;1 cDNA. In order to improve probe penetration into the tissue, the probe was partially degraded to an average length of 150 nucleotides. Sections were prepared for hybridization according to Scheres et al. (1990) and hybridized overnight at 42°C in 50% (v/v) formamide, 300 mM NaCl, 10 mM Tris–HCl pH 7.5, 1 mM EDTA, 0.02% (w/v) Ficoll, 0.02% (w/v) polyvinylpyrrolidone, 0.025% (w/v) bovine serum albumin (BSA), 10% (v/v) dextran sulfate and 60 mM DTT. After hybridization the sections were treated with a solution containing 500 mM NaCl, 1 mM EDTA, 10 mM Tris–HCl and 50 μg/ml RNase A. Finally, sections were washed several times in a 2xSSC solution. Hybridization signals were visualized with anti-digoxigenin antibodies conjugated with alkaline phosphatase. Images were processed using Photoshop 6 software (Adobe Systems Inc., San Jose, CA, USA).
Visualization of GFP
The GFP expression was analyzed using a Leica TCS SP2-AOBS confocal microscope. Green fluorescence of AMT1;1-GFP was excited at 488 nm with an argon laser. Emission was detected with a spectral detector set between 505 and 530 nm. To induce plasmolysis, whole roots were submerged in 500 mM mannitol solution for 30 min and then mounted on microscope slides in the same solution.
Acetylene reduction activity (ARA)
For the measurement of ARA of nodulated roots, plants were harvested and the root system was immediately detached and incubated at 25°C in 15 ml rubber-cap tubes containing 1/10 (vol/vol) acetylene. The ethylene produced at different time points was quantified with a sigma 3B gas chromatograph (Perkin-Elmer, Foster, CA, USA) equipped with a Porapak T column.
Results
Generation and molecular characterization of transgenic LjAMT1;1-antisense lines
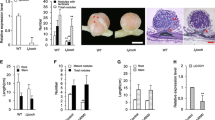
We generated transgenic L. japonicus GIFU plants carrying an antisense sequence that covered the 1,509 bp of the LjAMT1;1 cDNA. The antisense sequence was expressed from the soybean (Glycine max) leghemoglobin promoter (LBC3), which is specifically active in the invaded cells of the central tissue in determinate nodules (Lauridsen et al. 1993), and was terminated by the 3′ nopaline synthase terminator (nosT). T-DNA carrying the antisense-construct was introduced into L. japonicus via A. tumefaciens-mediated transformation and primary transformants (T0) were selected on the basis of G418 resistance and confirmed by PCR detection of the transgene in isolated genomic DNA (data not shown). Twelve LjAMT1;1-antisense primary transformant plants were transferred to the greenhouse and eight lines were characterized in subsequent generations. AMT1;1 transcript abundance in root nodules of G418-resistant T1 plants was measured by real-time RT-PCR on total RNA extracted from mature nodules (21 days post-inoculation, d.p.i.). Three out of eight antisense lines exhibited significantly-lower LjAMT1;1 transcript levels than the control (Fig. 1a). More specifically, lines Lj125, Lj126 and Lj131 had 25%, 50% and 60% lower AMT1;1 transcript levels than wild type plants, respectively. No changes in steady-state LjAMT1;1 transcript levels in roots and leaves were observed in these lines relative to the control (Fig. 1b, c), confirming the organ-specificity of the antisense effect. We speculated that much of the remaining LjAMT1;1 transcript in nodules of LjAMT1;1 antisense plants may have been confined to the vascular tissues (D’Apuzzo et al. 2004) which would not be expected to be affected by the antisense sequence driven by the LBC3 promoter. To test this idea, we carried out in situ RNA-hybridization analysis to visualize and compare cellular localization of the LjAMT1;1 transcript in wild type, Lj126 and Lj131 antisense nodules. LjAMT1;1 transcript was detected in both the central infected zone and vascular tissues of 3 weeks old nodules on wild type plants (Fig. 2a). In contrast, LjAMT1;1 transcript levels were clearly reduced in the central, infected zone but not in vascular bundles of antisense lines Lj126 and Lj131 (Fig. 2b, c). No significant hybridization signal above background was detected in wild type nodules hybridized to sense LjAMT1;1 probe (Fig. 2d).
Reduction of LjAMT1;1 transcript levels in nodules of antisense lines. Relative LjAMT1;1 transcript levels in nodules (a), roots (b), and leaves (c) 3 weeks after inoculation. Transcript levels were determined by qRT-PCR, normalized to that of the internal control ubiquitin (see Material and methods) and plotted relative to LjAMT1;1 transcript levels in wild type plants. Bars represent the mean and standard deviation of data obtained with RNA extracted from two biological replicates and three technical replicates each
In situ localization of LjAMT1;1 transcript in nodules of wild-type and LjAMT1;1 antisense plants. Longitudinal 8-μm thick sections of 3 weeks old nodules were hybridized to a 300 bp 3′-end LjAMT1;1 antisense and sense probes. Hybridization signal was visible as blue-purple precipitate. (a–c) hybridization with antisense probe, (d) hybridization with sense probe. (a) Bright-field image of a wild type nodule. (b) Bright-field image of a Lj126 antisense line nodule. (c) Bright-field image of a Lj131 antisense line nodule. (d) Bright-field image of a wild type nodule. Arrowheads indicate signal detected in vascular bundles
Reduction of LjAMT1;1 transcript level in the infected zone of Lotus nodules leads to increased nodulation
T1 seeds of Lj125, Lj126 and Lj131 antisense lines showed a 3:1 Mendelian segregation of the G418-resistance phenotype (Table 1), suggesting a single T-DNA integration event in each line. To determine the physiological impact of reduced AMT1;1 expression in the nitrogen-fixing zone of Lotus nodules, nodulation capacities of wild type and antisense plants were compared. Seedlings were inoculated with M. loti NZP2133 6 days after sowing (Barbulova and Chiurazzi 2005) and the kinetics of nodulation were followed for 6 weeks (Fig. 3). Initial experiments were carried out with a segregating population of T1 plants without G418-selection, to avoid any negative effects of the antibiotic on nodulation. Approximately three-quarters of all plants of the segregating lines Lj125, Lj126 and Lj131 produced significantly more nodules than wild type, consistent with a 3:1 segregation ratio of a single T-DNA insert in the T1 generation (Fig. 3a–c; Table 1). Analysis of the kinetics of nodulation revealed a substantial increase in nodule numbers on antisense plants compared to the wild-type, between 2 and 3 weeks after inoculation (Fig. 3d–f).
Nodulation kinetics of wild-type and LjAMT1;1 antisense plants. The number of nodules per plant was counted for T1 segregants of LjAMT1;1 antisense lines Lj125 (a, 1–25), Lj126 (b, 1–18), Lj131 (c, 1–18), the wild-type (black bars) and a transgenic line used as a control (grey bars). Plants were checked at 6 weeks post inoculation (7 weeks after sowing) and only mature nodules were scored. Kinetics of nodulation for five Lj125 (d), Lj126 (e) and Lj131 (f) T1 segregants and wild type plants. Wild-type data represent the mean and standard deviations obtained from three independent experiments (16 plants per experiment)
The enhanced-nodulation phenotype of LjAMT1;1 antisense lines was confirmed on T2 plants first selected for G418-resistance and then inoculated with M. loti. Lines Lj126 and Lj131 developed approximately twice the number of nodules as a G418-resistant, transgenic plant used as control (Fig. 4). Antisense plants continued to form new nodules on young roots 2–3 weeks after inoculation, while wild-type plants did not (Fig. 5a).
Nodulation phenotype of LjAMT1;1 antisense and wild-type plants infected with wild-type or nifH− mutant rhizobia. G418-resistant LjAMT1;1 antisense and transgenic controls were inoculated 1 week after germination with either wild-type or a nifH − strain of M. loti. Nodule numbers were analyzed 5 weeks post inoculation. Control transgenic line was infected with wild-type (black bar) and nifH − rhizobia (grey bar), while Lj126 and Lj131 antisense lines were infected with wild-type M. loti (white bars). The data represent mean and standard deviation obtained from three independent experiments (16 plants per experiment)
Nodulation pattern and nitrogen fixation in LjAMT1;1 lines. The pattern of nodulation in transgenic G418-resistant control (a, left panel) and Lj126 plants (a, right panel) was scored at 5 weeks post inoculation. Younger nodules observed on the secondary roots are shown in the square. (b) Acetylene reduction activities per nodule of G418-resistant transgenic control (black bar), and Lj125, Lj126, Lj131 plants (white bars). (c) Nodule fresh weight of G418-resistant transgenic control (black bar), and Lj125, Lj126, Lj131 plants (white bars). (d) Shoots fresh weight (mg/plant) of G418-resistant transgenic control infected with wild-type (black bar) or nifH − rhizobia (grey bar) and Lj125, Lj126, Lj131 antisense lines infected with M. loti wild-type (white bars). Data are mean and SD of 32 plants from two independent experiments
Nitrogen-fixing capacity of LjAMT1;1 antisense nodules
In view of the enhanced nodulation of LjAMT1;1 antisense plants, we were interested in testing the nitrogen-fixing activity of these plants. Using the acetylene reduction assay to estimate nitrogenase activity, we found that the nitrogen fixation activity of each nodule of these plants was significantly lower (40–45%) than that of wild-type nodules (Fig. 5b). The analysis of the nodule biomass (Fig. 5c) indicated that the nitrogen-fixation reduction found in the nodules of antisense lines was not due to a reduction of the nodules size. Interestingly, the growth of two antisense lines under symbiotic nitrogen-fixing conditions actually exceeded that of wild type plants (Fig. 5d), suggesting that the total amount of nitrogen fixed over the entire growth period was comparable or greater in the anti-sense plants.
LjAMT1;1 expression is under developmental but not nitrogen-control in nodules
We utilized an AMT1;1 promoter-gusA fusion (D’Apuzzo et al. 2004) to monitor LjAMT1;1 expression in transgenic plants after infection with M. loti wild-type or a nifH − (fix −) mutant strain. After infection of Lotus wild-type plants with the nifH − strain, an increased number of white nodules can be detected at 5 weeks post inoculation (Fig. 4). Despite the lack of nitrogen fixation, the pattern and intensity of GUS activity in young nodules was similar in nodules elicited by wild-type and nifH − M. loti (Fig. 6). In other words, although we cannot exclude the existence of a posttranscriptional mechanism of LjAMT1;1 mRNA regulation in nodules (Yuang et al. 2007a), its nodular expression seems not to be dependent by the presence of ammonium, the product of nitrogen fixation and the substrate of the LjAMT1;1 transporter.
Localization of LjAMT1;1-GFP fusion protein to the PM
To investigate the subcellular localization of LjAMT1;1 protein in the root tissues of Lotus plants, we constructed a translational fusion of the AMT1;1 gene to the green fluorescent protein (GFP) gene. GFP was fused in frame at the N-terminal end of AMT1;1. The coding sequence for the fusion protein was placed downstream of the CAMV-35S promoter and introduced into Lotus by A. rhizogenes-mediated transformation. GFP fluorescence was observed in the transgenic hairy root cells using a confocal laser-scanning microscope. GFP fluorescence was confined to the periphery of cortical root cells (Fig. 7a) and after plasmolysis, fluorescence was retained on the detached PM (Fig. 7b, c). The same PM localization has been reported for AMT proteins in other plant species (Ludewig et al. 2003; Simon-Rosin et al. 2003; Yuan et al. 2007b).
Plasma membrane localization of AMT1;1-GFP in transgenic roots. The sub-cellular localization of AMT1;1-GFP was examined in transgenic hairy roots. (a) GFP fluorescence in transgenic cortical root cells. (b) Nomarsky DIC visualization of transgenic root cells after plasmolysis in 0.5 M mannitol. (c) DIC/GFP overlay
Discussion
We used a reverse-genetic approach to investigate the role played by LjAMT1;1, in Lotus nodules. Expression of an antisense version of LjAMT1;1 in stably-transformed plants was driven by the GmLBC3 promoter, which is specifically active in infected cells of the nitrogen fixation zone of determinate nodules (Lauridsen et al. 1993). Three independent transgenic lines with reduced expression of LjAMT1;1 in nodules were obtained (Fig. 1). Down-regulation of AMT1;1 appeared to be localized in the central region of the nodules and no spreading of RNA silencing was observed in the surrounding vascular tissues (Fig. 2). Recently, Complainville et al. (2003) described changes in the pattern of symplastic communication observed during nodule development, by analyzing the un-loading and distribution of GFP fluorescence in transgenic M. truncatula plants where GFP expression was driven by the A. thaliana companion cell-specific AtSUC2 promoter (Imlau et al. 1999). In particular, GFP fluorescence was present in the meristematic and invasion zones of mature nodules, whereas no GFP fluorescence was detected in cells of the central, infected zone, suggesting a reduction in the permeability of plasmodesmata in that region (Complainville et al. 2003). A reduction in symplastic connectivity between cells of the infected zone and vascular tissues of nodules could explain the absence of spreading of RNA silencing from the former to the latter in the LjAMT1;1 antisense lines (Fig. 2).
Intriguingly, antisense inhibition of LjAMT1;1 led to an increase in nodule number in the three silenced transgenic lines (Figs. 3, 4). The number of nodules on the silenced lines was 2–3 times higher than on wild type and transgenic control roots (Fig. 3, 4).
Nodulation and nitrogen fixation levy a substantial metabolic cost on plants. Legumes employ a number of regulatory mechanisms to avoid nodulation under N-replete condition, when sufficient mineral or organic nitrogen is available in the soil, and to restrict nodulation to sustainable levels when soil nitrogen is limiting. The number of nodules is regulated in at least two ways: by aborting infection and by confining the nodulation zone. Defects in these mechanisms, caused by genetic mutation lead to hyper- or super-nodulation (Carroll et al. 1985; Caetano-Anollès and Gresshoff 1991; Penmetsa and Cook 1997; Nishimura et al. 2002; Krusell et al. 2002; Oka-Kira et al. 2005). On the other hand, a strict interdependence exists between the level of nitrogenase activity and the efficiency of carbon sources flux providing energy for the bacteroid-mediated N2 reduction (Pathirana et al. 1992; Schulze et al. 1998). Thus, negative feedback mechanisms governing nodulation, respond to both the number of nodules already initiated and the nitrogen-fixation output of mature nodules, although how plants monitor these parameters remains poorly understood. It appears that plants are able to compensate for inefficient nodules, caused either by plant and/or bacteria genetic lesions, at least to some extent, by increasing nodule number, although this does little good when mutations completely abolish nitrogen fixation (Hirsch and Smith 1987; Gordon et al. 1999; Suganuma et al. 2003). The pattern of nodulation of the LjAMT1;1 antisense lines, with a low density of nodules distributed in a wider zone of the root system (Fig. 5a) as well as their nodulation kinetics (Fig. 3d–f) are reminiscent of the phenotype exhibited by many of the nitrogen fixation-impaired plant mutants, which exhibit normal nodulation kinetics up to the time when nitrogen fixation usually begins but enhanced nodulation compared to the wild-type afterwards (Cordoba et al. 2003; Gordon et al. 1999; Suganuma et al. 2003; Krusell et al. 2005).
Interestingly, it appears that the reduced ARA activity detected in the antisense nodules (Fig. 5b), that was not associated to a reduced biomass (Fig. 5c), was compensated for by increased nodulation, since shoots of LjAMT1;1 antisense were at least as large as those of control plants (Fig. 5d).
The analysis of the LjAMT1;1 antisense lines described in this paper, provides new insight into the symbiotic role played by high affinity ammonium transporters. An involvement of AMT1;1 in the transport of ammonium across the SM is unlikely since the biochemical properties of AMT1;1 (K m = 1.7 μM; D’Apuzzo et al. 2004), is not compatible with such a role, on the basis of the ammonium concentrations estimated within the symbiosome space (12 mM; Streeter 1989). Besides, no AMT proteins have been identified on purified PBM (Wienkoop and Saalbach 2003; Catalano et al. 2004). We have shown a PM location of the LjAMT1;1 protein (Fig. 7), a result identical to that obtained for different AMT1 proteins in Lotus and other plant species (Ludewig et al. 2003; Simon-Rosin et al. 2003; Yuan et al. 2007b). It has been proposed that PM-located AMT1 proteins may play a role in recovery of ammonium lost from cells via diffusion, prior to its incorporation into amino acids (Simon-Rosin et al. 2003). The spatial expression pattern of LjAMT1;1 in nodules (Fig. 6) and the putative PM location of the protein are consistent with such a role in Lotus. However, we cannot exclude a possible posttranscriptional regulation of the LjAMT1;1 mRNA in nodules (Yuang et al. 2007a). This action would not be dependent by nitrogen fixation, but rather associated to the nodule developmental program (Fig. 6). Inefficient retrieval of ammonium lost from nitrogen-fixing cells in the infected zone of LjAMT1;1 anti-sense nodules would be expected to reduce the efficiency of such nodules, and could account for the apparent attenuation of feedback inhibition and consequent increase in nodule number on the antisense plants. Possibly, the reduced ammonium availability, could affect the nitrogen-fixation activity observed in the nodules of antisense plants (Fig. 5b), by altering the rate of amino-acids synthesis and cycling that was proposed to drive N2 fixation (Lodwig et al. 2003).
An intriguing, alternative hypothesis, is that LjAMT1;1 could play a role as a direct sensor of ammonium, or by transducing the signal by the actual sensor; identifying a putative late check-point of the nodules functionality. A sensing function has been proposed for fungal Amt high affinity transporters (Javelle et al. 2003) and for the Saccharomyces cerevisiae Mep2 protein that links low ammonium availability with a developmental program represented by pseudohyphal differentiation (Lorenz and Heitman 1998).
References
Barbulova A, Chiurazzi M (2005) Procedure for Lotus japonicus in vitro nodulation studies. In: Marquez AJ (ed) Lotus japonicus handbook. Springer, Dordrecht, pp 83–86
Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20:1195–1197. doi:10.1007/BF00028908
Beringer JE (1974) R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84:188–198
Caetano-Anollès G, Gresshoff PM (1991) Plant genetic control of nodulation. Annu Rev Microbiol 45:345–382. doi:10.1146/annurev.mi.45.100191.002021
Carroll BJ, McNeil DL, Gresshoff PM (1985) A supernodulation and nitrate-tolerant symbiotic (nts) soybean mutant. Plant Physiol 78:34–40
Catalano CM, Lane WS, Sherrier AJ (2004) Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25:519–531. doi:10.1002/elps.200305711
Complainville A, Brocard L, Roberts I, Dax E, Sever N, Sauer N et al (2003) Nodule initiation involves the creation of a new symplasmic field in specific root cells of Medicago species. Plant Cell 120:2778–2791. doi:10.1105/tpc.017020
Cordoba E, Shishkova S, Vance CP, Hernandez G (2003) Antisense inhibition of NADH glutamate synthase impairs carbon/nitrogen assimilation in nodules of alfalfa (Medicago sativa L.). Plant J 33:1037–1049. doi:10.1046/j.1365-313X.2003.01686.x
Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M (2007) The expanded family of ammonium transporters in the perennial poplar plant. New Phytol 174:137–150. doi:10.1111/j.1469-8137.2007.01992.x
D’Apuzzo E, Rogato A, Simon-Rosin U, El Alaoui H, Barbulova A, Betti M et al (2004) Characterisation of three functional high affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol 134:1763–1774. doi:10.1104/pp.103.034322
Duby G, Oufattole M, Boutry M (2001) Hydrophobic residues within the predicted N-terminal amphiphilic a-helix of a plant mitochondrial targeting presequence play a major role in in vivo import. Plant J 27:539–549. doi:10.1046/j.1365-313X.2001.01098.x
Flemetakis E, Efrose RC, Desbrosses G, Dimou M, Delis C, Aivalakis G et al (2004) Induction and spatial organization of polyamine biosynthesis during nodule development in Lotus japonicus. Mol Plant Microbe Interact 17:1283–1293. doi:10.1094/MPMI.2004.17.12.1283
Gordon AJ, Minchin FR, James CL, Komina O (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120:867–877. doi:10.1104/pp.120.3.867
Handberg K, Stougaard J (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2:487–496. doi:10.1111/j.1365-313X.1992.00487.x
Hirsch AM, Smith CA (1987) Effect of Rhizobium meliloti nif and fix mutants on alfalfa root nodule development. J Bacteriol 169:1137–1146
Howitt SM, Udvardi MK (2000) Structure, function and regulation of ammonium transporters in plants. Biochim Biophys Acta 1465:152–170. doi:10.1016/S0005-2736(00)00136-X
Howitt SM, Udvardi MK, Day DA, Gresshoff PM (1986) Ammonia transport in free-living and symbiotic Rhizobium sp. ANU289. Microbiol 132:257–261
Husted S, Schjoerring JK (1996) Ammonia flux between oilseed rape plants and the atmosphere in response to changes in leaf temperature, light intensity, and hair humidity (interactions with leaf conductance and apoplastic NH4 + and H+ concentrations). Plant Physiol 112:67–74
Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11:309–322
Javelle A, Andre B, Marini AM, Chalot M (2003) High-affinity ammonium transporters and nitrogen sensing in mycorrhizas. Trends Microbiol 11:53–55. doi:10.1016/S0966-842X(02)00012-4
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405. doi:10.1007/BF02667740
Jensen HL (1942) Nitrogen fixation in leguminous plants. General characters of root-nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc Linn Soc Lond 66:98–108
Kistner C, Matamoros M (2005) RNA isolation using phase extraction and LiCl precipitation. In: Marquez AJ (ed) Lotus japonicus handbook. Springer, Dordrecht, pp 123–124
Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K et al (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420:422–426. doi:10.1038/nature01207
Krusell L, Krause K, Ott T, Desbrosses G, Kramer U, Sato S et al (2005) The Sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17:1625–1636. doi:10.1105/tpc.104.030106
Lauridsen P, Franssen H, Stougaard J, Bisseling T, Marcker KA (1993) Conserved regulation of the soybean early nodulin ENOD2 gene promoter in determinate and indeterminate transgenic root nodules. Plant J 3:483–492. doi:10.1046/j.1365-313X.1993.t01-25-00999.x
Lodwig EM, Hosle AHF, Bourdès A, Findlay K, Allaway D, Karunakaran R et al (2003) Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422:722–726. doi:10.1038/nature01527
Lorenz MC, Heitman J (1998) Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150:1443–1457
Ludewig U, von Wiren N, Frommer WB (2002) Uniport of NH4 + by the root hair plasma membrane ammonium transporter LeAMT1;1. J Biol Chem 277:13548–13555. doi:10.1074/jbc.M200739200
Ludewig U, Wilken S, Wu B, Jost W, Obrdlik P, El Bakkoury M et al (2003) Homo- and hetero-oligomerization of ammonium transporter-1 NH4 uniporters. J Biol Chem 278:45603–45610. doi:10.1074/jbc.M307424200
Martirani L, Stiller J, Mirabella R, Alfano F, Lamberti A, Radutoiu SE et al (1999) Establishment of a T-DNA tagging program in the model legume Lotus Japonicus. Expression patterns, activation frequencies and potential for insertional mutagenesis. Mol Plant Microbe Interact 12:275–284. doi:10.1094/MPMI.1999.12.4.275
Niemietz CM, Tyerman SD (2000) Channel-mediated permeation of ammonia gas through the peribacteroid membrane of soybean nodules. FEBS Lett 465:110–114. doi:10.1016/S0014-5793(99)01729-9
Nishimura R, Ohmori M, Kawaguchi M (2002) The novel symbiotic phenotype of enhanced-nodulation mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol 43:853–859. doi:10.1093/pcp/pcf098
Oka-kira E, Tateno K, Miura K, Haga T, Hayashi M, Harada K et al (2005) Klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J 44:505–515. doi:10.1111/j.1365-313X.2005.02543.x
Pathirana AM, Vance CP, Miller SS, Gantt S (1992) Alfalfa root nodule phosphoenolpiruvate carboxylase: characterization of the cDNA and expression in effective and plant-controlled ineffective nodules. Plant Mol Biol 20:437–450. doi:10.1007/BF00040603
Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275:527–530. doi:10.1126/science.275.5299.527
Roberts DM, Tyerman SD (2002) Voltage-dependent cation channels permeable to NH(+)(4), K(+), and Ca(2+) in the symbiosome membrane of the model legume Lotus japonicus. Plant Physiol 128:370–378. doi:10.1104/pp.128.2.370
Salvemini F, Marini AM, Riccio A, Patriarca EJ, Chiurazzi M (2001) Functional characterisation of an ammonium transporter gene from Lotus japonicus. Gene 270:237–243. doi:10.1016/S0378-1119(01)00470-X
Scheres B, van de Wiel C, Zalensky A, Horvath B, Spaink HP, van Eck H et al (1990) The ENOD12 gene product is involved in the infection process during the pea–Rhizobium interaction. Cell 60:281–294. doi:10.1016/0092-8674(90)90743-X
Schulze J, Shi L, Blumenthal J, Samac DA, Gantt JS, Vance JP (1998) Inhibition of alfalfa root nodule phosphoenolpyruvate carboxylase through an antisense strategy impacts nitrogen fixation and plant growth. Phytochemistry 49:341–346. doi:10.1016/S0031-9422(98)00221-0
Simon-Rosin U, Wood C, Udvardi MK (2003) Molecular and cellular characterisation of LjAMT2;1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol Biol 51:99–108. doi:10.1023/A:1020710222298
Sohlenkamp C, Shelden M, Howitt S, Udvardi M (2000) Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett 467:273–278. doi:10.1016/S0014-5793(00)01153-4
Sonoda Y, Ikeda A, Saiki S, von Wiren N, Yamaya T, Yamaguchi J (2003) Distinct expression and function of three ammonium transporter genes (OsAMT1;1–1;3) in rice. Plant Cell Physiol 44:726–734. doi:10.1093/pcp/pcg083
Streeter J (1989) Estimation of ammonium concentration in the cytosol of soybean nodules. Plant Physiol 90:779–782
Suenaga A, Moriya K, Sonoda Y, Ikeda A, von Wiren N, Hayakawa T et al (2003) Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol 44:206–211. doi:10.1093/pcp/pcg017
Suganuma N, Nakamura Y, Yamamoto M, Ohta T, Koiwa H, Akao S et al (2003) The Lotus japonicus Sen1 gene controls rhizobial differentiation into nitrogen-fixing bacteroids in nodules. Mol Gen Genom 269:312–320. doi:10.1007/s00438-003-0840-4
Udvardi MK, Day DA (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48:493–523. doi:10.1146/annurev.arplant.48.1.493
Von Wiren N, Merrick M (2004) Regulation and function of ammonium carriers in bacteria, fungi and plants. Topics in current genetics. In: Boles E, Kramer R (eds) Molecular mechanisms controlling transmembrane transport. Springer, Berlin, pp 1–26
Wienkoop S, Saalbach G (2003) Proteome analysis. Novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol 131:1080–1090. doi:10.1104/pp.102.015362
Wood CC, Porèe F, Dreyer I, Koehler GJ, Udvardi MK (2006) Mechanisms of ammonium transport, accumulation, and retention in ooyctes and yeast cells expressing Arabidopsis AtAMT1;1. FEBS Lett 580:3931–3936. doi:10.1016/j.febslet.2006.06.026
Yuan L, Loquè D, Ye F, Frommer WB, von Wiren N (2007a) Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiol 143:732–744. doi:10.1104/pp.106.093237
Yuan L, Loquè D, Kojima S, Rauch S, Ishiyama K, Inoue E et al (2007b) The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19:2636–2652. doi:10.1105/tpc.107.052134
Acknowledgements
We wish to thank C. Ronson and Mette Groenlund for providing the M. loti nifH − strain and the pGPTVkan-lbc3 plasmid, respectively. We thank Nunzia Bellopede, Anna Sollo and Chiara Lepore for technical assistance. We also thank Biagio Giordano and the gardeners of the Royal Botanical Garden of Naples for their excellent plant care. This work was supported by a grant from the EEC (INTEGRAL: MRTN-CT-2003-505227). S.O. was supported by an EEC fellowship (INTEGRAL: MRTN-CT-2003-505227).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rogato, A., D’Apuzzo, E., Barbulova, A. et al. Tissue-specific down-regulation of LjAMT1;1 compromises nodule function and enhances nodulation in Lotus japonicus . Plant Mol Biol 68, 585–595 (2008). https://doi.org/10.1007/s11103-008-9394-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9394-5