Abstract
Some plants like Arabidopsis thaliana increase in freezing tolerance when exposed to low nonfreezing temperatures, a process known as cold acclimation. Other plants including tomato, Solanum lycopersicum, are chilling sensitive and incur injury during prolonged low temperature exposure. A key initial event that occurs upon low temperature exposure is the induction of genes encoding the CBF transcription factors. In Arabidopsis three CBF genes, present in a tandemly-linked cluster, are induced by low temperatures. Tomato also harbors three tandemly-linked CBF genes, Sl-CBF3–CBF1–CBF2, but only one of these, Sl-CBF1, is low-temperature responsive. Here we report that Solanum species that are closely-allied to cultivated tomato essentially share this structural organization, but the locus is in a dynamic state of flux. Additional paralogs and in-frame deletions between adjacent genes occur, and the genomic regions flanking the CBF genes are dissimilar across Solanum species. Nevertheless, the CBF1 upstream region remains intact and highly conserved. This feature differed for CBF2 and CBF3, whose upstream regions were far less conserved. CBF1 was also the only low-temperature responsive gene in the cluster and its expression was greatly affected by a circadian clock. The tuber-bearing S. tuberosum and S. commersonii also harbored a fourth gene, CBF4, which was also low temperature responsive. CBF4 was physically linked to CBF5 in S. tuberosum, but CBF5 was absent from S. commersonii. Phylogenic analyses suggest that CBF5–CBF4 resulted from the duplication of the CBF3–CBF1–CBF2 cluster. DNA sequence motifs shared between the Solanum CBF1 and CBF4 upstream regions were identified, portions of which were also present in the Arabidopsis CBF1-3 upstream regions. These results suggest that much greater functional constraints are placed upon the Solanum CBF1 upstream regions over the other CBF upstream regions and that CBF4 has retained the capacity for low temperature responsiveness following the duplication event that gave rise to CBF4.







Similar content being viewed by others
Abbreviations
- CBF:
-
C-repeat binding factor
- CBF/DREB1:
-
C-repeat binding factor/dehydration responsive element binding 1
- CDS:
-
Coding sequence
- CRT/DRE:
-
C-repeat/dehydration responsive element
- HC:
-
Hydrophobic cluster
- ICEr:
-
Induction of CBF expression region
- ZT:
-
Zeitgeber time
References
Alexandrov NN, Troukhan ME, Brover VV, Tatarinova T, Flavell RB, Feldmann KA (2006) Features of Arabidopsis genes and genome discovered using full-length cDNAs. Plant Mol Biol 60:69–85
Alonso-Blanco C, Gomez-Mena C, Llorente F, Koornneef M, Salinas J, Martinez-Zapater JM (2005) Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol 139:1304–1312
Badawi M, Danyluk J, Boucho B, Houde M, Sarhan F (2007) The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Mol Genet Genomics 277:533–554
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942
Chen HH, Li PH (1980) Characteristics of cold acclimation and deacclimation in tuber-bearing Solanum species. Plant Physiol 65:1146–1148
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
D’Arcy WG (1991) The Solanaceae since 1976 with a review of its biogeography. In: Hawkes JG, Lester RN, Nee M Estrada N (eds) Solanaceae III. Royal Botanic Gardens/Kew, London, pp 75–138
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Fowler SG, Cook D, Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137:961–968
Fujita M, Umemura M, Yoko-o T, Jigami Y (2006) PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol Biol Cell 17:5253–5264
Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16:433–442
Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54:767–781
Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648
Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110–2113
Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT (2002) Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 129:1086–1094
Hughes JD, Estep PW, Tavazoie S, Church GM (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296:1205–1214
Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127:910–917
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyere C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pe ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quetier F, Wincker P (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004) dwarf and delayed-flowering 1, A novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 37:720–729
Marsan L, Sagot MF (2000) Algorithms for extracting structured motifs using a suffix tree with an application to promoter and regulatory site consensus identification. J Comput Biol 7:345–362
Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8:122–128
Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. EMBNET NEWS 1–4
Novillo F, Medina J, Salinas J (2007) Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci USA 104:21002–21007
Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, Ikeo K, Itoh T, Gojobori T, Sasaki T (2006) The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res 34:D741–D744
Pavesi G, Mereghetti P, Mauri G, Pesole G (2004) Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res 32:W199–W203
Pavesi G, Zambelli F, Pesole G (2007) WeederH: an algorithm for finding conserved regulatory motifs and regions in homologous sequences. BMC Bioinformatics 8:46
Pennycooke JC, Cheng H, Stockinger EJ (2008) Comparative genomic sequence and expression analyses of Medicago truncatula and M. sativa ssp. falcata cold-acclimation specific (CAS) genes. Plant Physiol 146:1–13
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10:577–586
Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, Muramatsu M, Hayashizaki Y, Kawai J, Carninci P, Itoh M, Ishii Y, Arakawa T, Shibata K, Shinagawa A, Shinozaki K (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296:141–145
Semple C, Wolfe KH (1999) Gene duplication and gene conversion in the Caenorhabditis elegans genome. J Mol Evol 48:555–564
Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun 250:161–170
Skinner JS, von Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen TH, Hayes PM (2005) Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol 59:533–551
Stockinger EJ, Walling LL (1994) Pto3 and Pto4: novel genes from Lycopersicon hirsutum var. glabratum that confer resistance to Pseudomonas syringae pv tomato. Theor Appl Genet 89:879–884
Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94:1035–1040
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thompson W, Rouchka EC, Lawrence CE (2003) Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res 31:3580–3585
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Dejardin A, Depamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M., Jorgensen R, Joshi C, Kangasjarvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leple JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouze P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Vallejos CE, Pearcy RW (1987) Differential acclimation potential to low temperature in two species of Lycopersicon: photosynthesis and growth. Can J Bot 65:1303–1307
Van Der Ploeg A, Heuvelink E (2005) Influence of sub-optimal temperature on tomato growth and yield: a review. J Hortic Sci Biotechnol 80:652–659
Venema JH, Posthumus F, van Hasselt PR (1999) Impact of suboptimal temperature on growth, photosynthesis, leaf pigments and carbohydrates of domestic and high-altitude wild Lycopersicon species. J Plant Physiol 155:711–718
Venema JH, Linger P, van Heusden AW, van Hasselt PR, Bruggemann W (2005) The inheritance of chilling tolerance in tomato (Lycopersicon spp.). Plant Biol (Stuttg) 7:118–130
Wang Z, Triezenberg SJ, Thomashow MF, Stockinger EJ (2005) Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functional redundancy in trans-activation. Plant Mol Biol 58:543–559
Wang X, Tang H, Bowers JE, Feltus FA, Paterson AH (2007) Extensive concerted evolution of rice paralogs and the road to regaining independence. Genetics 177:1753–1763
Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133:910–918
Zhang X, Fowler SG, Cheng H, Lou Y, Rhee SY, Stockinger EJ, Thomashow MF (2004) Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J 39:905–919
Acknowledgements
We thank David M. Francis, Esther van der Knaap, David E. Somers and Sophien Kamoun for helpful suggestions during the preparation of the manuscript. This work was supported by grants from the National Science Foundation Plant Genome Project (grant no. 0110124). Salaries and research support were also provided in part by state and federal funds appropriated to the Ohio Agricultural Research and development Center, The Ohio State University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Solanum CBF polypeptides in BOXSHADE-formatted alignment. The green, fuchsia, and blue boxes identify the CBF1 SER (S), the CBF2 ARG (R), and the CBF3 ARG-ALA-ASN-HIS-SER (RANHS) blocks, respectively. The orange box at the very NH3-termini of CBF polypeptides identifies the amino acid region shared between the CBF1 orthologs and the potato-clade CBF3, and tomato-clade CBF2 orthologs. The turquoise box identifies the abbreviated NH3-termini common to both CBF4 and CBF5 polypeptides. The violet box inclusive of and proximal to HC2 identifies amino acid residues shared between the tomato-clade CBF1 and CBF2 polypeptides that are distinct from the potato-clade CBF1 and CBF2, whereas the lime box between the E-rich motif and HC2 identifies additional residues present in- and shared across the potato-clade CBF1 and CBF2 polypeptides (and also with St-CBF4) that absent from the tomato-clade CBF1 and CBF2 polypeptides. Asterisks above the alignment identify shared residues between CBF3 and CBF5 polypeptides that are distinct from the other CBF polypeptides. The downward black arrow identifies the AP2 domain ARG residue in all tomato-clade CBFs that is a MET residue in all potato-clade CBFs. The downward red arrow identifies the junction point of the Sp-CBF1/2 fusion (DOC 73.5 kb)
Fig. S2
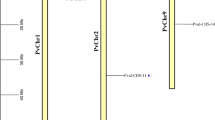
DNA blot hybridizations of S. commersonii (cmm) and S. tuberosum (tbr) DNAs with gene-specific CBF probes. Hybridization probes are indicated above the lanes. B = Bgl I, E = Eco RI, V = Eco RV, X = Xba I. (The additional fragment cross-hybridizing with CBF2 in the Xba I lane results from an Xba I site dividing the region used as probe (TIF 1404 kb)
Fig. S3
Solanum CBF upstream regions in BOXSHADE-formatted sequence alignment. S3A, Alignment of the Solanum CBF1 ortholog upstream regions and portions of the CBF4 upstream regions sharing identity with the CBF1 gene regions. CBF1 sequences begin at the termination codon of the upstream CBF3 gene and continue to the AUG initiator codon (denoted as +1). S. tuberosum CBF4 upstream nucleotides −2048 to −3333 are included in the alignment (i.e., nucleotides −1 to −2048 upstream of CBF4 are omitted). Similarly, only S. commersonii upstream nucleotides −2366 to −2170 are included in the alignment. S3B, Solanum CBF2 upstream regions. S3C, Solanum CBF3 upstream regions. S3D, Alignment of the Sc-CBF4 and St-CBF4 upstream regions. Boxed regions are described in the text (DOC 576 kb)
Rights and permissions
About this article
Cite this article
Pennycooke, J.C., Cheng, H., Roberts, S.M. et al. The low temperature-responsive, Solanum CBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Mol Biol 67, 483–497 (2008). https://doi.org/10.1007/s11103-008-9333-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9333-5