Abstract
SQUAMOSA PROMOTER BINDING PROTEIN-LIKEs (SPLs) encode plant-specific transcription factors that regulate plant growth and development, stress response, and metabolite accumulation. However, there is limited information on Scutellaria baicalensis SPLs. In this study, 14 SbSPLs were identified and divided into 8 groups based on phylogenetic relationships. SbSPLs in the same group had similar structures. Abscisic acid-responsive (ABRE) and MYB binding site (MBS) cis-acting elements were found in the promoters of 8 and 6 SbSPLs. Segmental duplications and transposable duplications were the main causes of SbSPL expansion. Expression analysis based on transcriptional profiling showed that SbSPL1, SbSPL10, and SbSPL13 were highly expressed in roots, stems, and flowers, respectively. Expression analysis based on quantitative real-time polymerase chain reaction (RT‒qPCR) showed that most SbSPLs responded to low temperature, drought, abscisic acid (ABA) and salicylic acid (SA), among which the expression levels of SbSPL7/9/10/12 were significantly upregulated in response to abiotic stress. These results indicate that SbSPLs are involved in the growth, development and stress response of S. baicalensis. In addition, 8 Sba-miR156/157 s were identified, and SbSPL1-5 was a potential target of Sba-miR156/157 s. The results of target gene prediction and coexpression analysis together indicated that SbSPLs may be involved in the regulation of L-phenylalanine (L-Phe), lignin and jasmonic acid (JA) biosynthesis. In summary, the identification and characterization of the SbSPL gene family lays the foundation for functional research and provides a reference for improved breeding of S. baicalensis stress resistance and quality traits.
Key message
This report details the characteristics and potential regulatory network of the S. baicalensis SbSPL gene family and demonstrates the potential role of SbSPLs in the abiotic stress response and material accumulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Scutellaria baicalensis is called Huang-Qin in Chinese, which means golden (precious) herb. It belongs to the perennial herb family Lamiaceae and is commonly used in traditional medicine. S. baicalensis is distributed in East Asia, Europe and North America, and it is used for adjuvant therapy of various diseases (Wen et al. 2022; Zhao et al. 2016). It has been officially listed in the Chinese Pharmacopoeia (2020), European Pharmacopoeia (EP 9.0) and British Pharmacopoeia (BP 2018). The root is the main medicinal part of S. baicalensis (Zhao et al. 2016), and flavonoids and their glycosides are the main medicinal components that have important antiviral (Zhao et al. 2019b), antidiabetic (Huang et al. 2017; Na and Lee 2019), and anticancer (Park et al. 2021; Xiang et al. 2022) functions. It is worth noting that the secondary metabolites of S. baicalensis have an important effect when used to treat coronavirus disease 2019 (COVID-19) (Malekmohammad and Rafieian-Kopaei 2021; Song et al. 2020). Liu et al. and Su et al. found that baicalein and wogonin in S. baicalensis extract show strong anti-COVID-19 activity in cell systems (Liu et al. 2021; Su et al. 2020). Therefore, research on S. baicalensis is of great medical importance.
Abiotic stress is a key factor affecting the yield and quality of S. baicalensis under natural conditions. First, abiotic stress produces reactive oxygen species (ROS) to damage the tissue cells of S. baicalensis. For example, Zhang et al. found that short-term drought led to an imbalance in ROS metabolism, and S. baicalensis regulated ROS levels by increasing the activities of protective enzymes such as superoxide dismutase (SOD) and peroxidase (POD) (Zhang et al. 2013). Second, the accumulation of secondary metabolites in S. baicalensis is associated with abiotic stress. Ultraviolet-B (UV-B) radiation can significantly affect the accumulation of secondary metabolite aglycones (including baicalein, wogonin and scutellarein) in tissue culture and produce oxidative stress (Yun et al. 2022). Yuan et al. found that abnormal temperature (10 °C and 40 °C) reduced the content of flavonoids in S. baicalensis, and high temperature affected the synthesis of two key enzymes, 7-O-glucuronosyltransferase and β-glucuronidase, in the interconversion of baicalin and baicalein (Yuan et al. 2012). Guo et al. also found that the content of secondary metabolites was positively correlated with temperature changes within a certain range (Guo et al. 2013). In addition, heavy metals such as cadmium, mercury and plumbum were also unfavorable factors for the growth and accumulation of medicinal components in S. baicalensis (Meng et al. 2022).
The transcription initiation process of eukaryotes is very complex and often requires the assistance of a variety of protein factors. Transcription factors (TFs) form a complex with RNA polymerase and participate in the transcription initiation process together. TFs in plants generally exist in the form of gene families and play an important role in plant signal transduction (Wani et al. 2021). SQUAMOSA-promoter binding protein-like (SPL) proteins are a family of plant-specific TFs found in many green plants. The first discovery of SPL TFs was in 1996, when Peter Huijser et al. cloned two genes in the Antirrhinum majus flower meristem. The genes both contain a conserved squamosa-promoter binding protein (SBP)-box domain and can bind to the promoter of the floral meristem characteristic gene SQUAMOSA (SQUA). The genes were later named AmSBP1 and AmSBP2 (Klein et al. 1996). Birkenbihl et al. found that the SBP-box protein is a highly conserved domain consisting of 76 amino acid residues and includes two finger-like zinc finger structures formed by the coordination of Zn2+, namely, Cys-Cys-Cys-His and Cys-Cys-His-Cys, and a nuclear localization signal (NLS) at the C-terminus of the SBP domain that partially overlaps the second zinc finger (Birkenbihl et al. 2005). In previous studies, members of the Arabidopsis thaliana AtSPL TFs were divided into 8 groups (I–VIII) (Cardon et al. 1999). With the development of plant whole-genome sequencing and bioinformatics, SPL TFs have been identified and analyzed in many species, including the dicotyledons Ziziphus jujuba (Shao et al. 2017), Fragaria vesca (Xiong et al. 2018), and Solanum lycopersicum (Salinas et al. 2012), and monocotyledons Oryza sativa (Xie et al. 2006), Triticum aestivum (Zhu et al. 2020), and Senna italica (Lai et al. 2022). An increasing number of studies have shown that SPL genes can actively respond to abiotic stresses such as drought, salt, and abnormal temperature. Overexpression of Vitis pseudoreticulata VpSBP16 could enhance the scavenging ability of ROS to enhance the salt tolerance and drought tolerance of grapes (Hou et al. 2018). In addition, 3 differentially expressed MsSPLs (MsSPL17, MsSPL23 and MsSPL36) were found in Medicago sativa under salt stress, which provided comprehensive information for the mechanism underlying salt tolerance improvement in M. sativa (He et al. 2022b).
MiR156/157 s are small noncoding RNAs (approximately 20 nt in length) produced by nuclease cutting and processing long-chain pri-miRNAs and are highly conserved in plants (Lopez-Ortiz et al. 2021). The sequences of miR156s and miR157s are highly similar, with only 1/2 different nucleotides (Zhou et al. 2021). Posttranscriptional mRNAs of SPLs with miRNA response elements (MREs) complementary to miR156 were cleaved and/or translationally repressed by miR156 (Rhoades et al. 2002). The key role of the miR156/157-SPL regulatory module in plant responses to abiotic stress has become increasingly clear. Eight of the 17 DgSPLs of Dactylis glomerata may be potential targets of miR156, and 2 of these 8 genes (DG1G01828.1 and DG0G01071.1) can simultaneously respond to drought, salt and heat stress (Feng et al. 2021). The tolerance of M. sativa to heat stress (40 °C) was increased after overexpression of miR156 and knockdown of MsSPL13 RNAi, suggesting that the miR156/SPL13 pathway contributes to the improvement of heat tolerance in M. sativa (Matthews et al. 2019). When A. thaliana was subjected to repeated heat stress, AtSPLs were posttranscriptionally downregulated by miR156, indicating that the miR156-SPL module mediated the A. thaliana response to repeated heat stress (Stief et al. 2014). In addition, SPL TFs can regulate plant physiological and metabolic processes to adapt to element-deficient environments. For example, microRNAs (Cu-miRNAs) can reduce the consumption of Cu by plants during periods of copper deficiency. Perea-García et al. found that Cu-deficiency responses mediated by AtSPL7 binding to Cu-miRNA promoters were often competitively inhibited by the miR156-SPL3 module in A. thaliana (Perea-García et al. 2021). In conclusion, previous studies have revealed the responsive function of miR156s-SPLs in a wide range of plant adaptive responses to the environment.
Although TFs have been widely studied in a large number of plants, the exploration of TFs in S. baicalensis, a precious Chinese herbal medicine, is rare. We searched Chinese and international journals and found only studies on WRKY TFs (Zhang et al. 2022a) and R2R3-MYB TFs in S. baicalensis (Wang et al. 2022). However, research on the SPL genes of S. baicalensis is completely lacking, and the upstream and downstream regulatory mechanisms of SbSPLs are also unknown. To this end, we screened SbSPLs and predicted and analyzed their gene structure, motifs, evolution, sequence conservation, cis-acting elements, expression patterns, and upstream and downstream regulatory relationships. The results of this study provide comprehensive information for understanding SbSPLs and provide clues for the interpretation of the S. baicalensis stress response and the regulation of metabolite biosynthesis.
Materials and methods
Identification and chromosomal localization of SbSPLs
The genome, protein and annotation files of S. baicalensis were downloaded from the National Genomics Data Center (https://ngdc.cncb.ac.cn/search/?dbId=gwh&q=PRJCA009554&page=1, Accessed on September 20, 2022) (Hu et al. 2022). Hidden Markov Model (HMM) seed files for SBP domains were downloaded from the Pfam database (http://pfam.sanger.ac.uk/, Accessed on 20 September 2022) (Punta et al. 2012). Hmmsearch was used to identify the SPL proteins of S. baicalensis, and the E-value threshold was 10–20 (Potter et al. 2018). Proteins with SBP domains were obtained through Pfam and SMART (http://smart.embl.de/, Accessed on September 20, 2022) database screening (Letunic and Bork 2018). The online ExPASy Bioinformatics Resource Portal (https://web.ExPASy.org/compute_pi/, Accessed on September 20, 2022) was used to predict the molecular weight (MW) and physicochemical properties of proteins (Duvaud et al. 2021). ProtComp 9.0 (http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc, Accessed on September 20, 2022) was used for subcellular localization prediction (Emanuelsson et al. 2000). TBtools was used to draw the chromosomal location map of SbSPLs based on the annotation files and genome files (Chen et al. 2020).
Sequence alignment and phylogenetic analysis
We aligned SbSPL and A. thaliana AtSPL protein sequences (UniProt database, https://www.UniProt.org) using MAFFT (Consortium 2017; Nakamura et al. 2018). The results are presented by group using Texmaker (Beitz 2000). Subsequently, a phylogenetic tree was constructed based on the alignment results using the maximum likelihood (ML) method in fasttree with a maximum of 1000 bootstrap replicates (Price et al. 2009). The R package ggtree was used for visualization (Yu et al. 2018).
Gene structure and conserved motif analysis
The location and length of the SbSPL CDS were clarified according to the annotation file. MEME Suite version 5.0.5 (http://meme.nbcr.net/meme/) was used to analyze conserved motifs in proteins (Bailey et al. 2009). The parameters were set as follows: maximum number of motifs = 10; optimum width of motifs = 5 to 50. The R package gggenes was used for the visualization of gene structure and conserved motifs (Gómez-Rubio 2017).
Prediction of cis-acting elements
Bedtools was used to extract the 2.0 kb promoter sequence upstream of SbSPLs (Quinlan 2014). Cis-acting elements were analyzed using the PLACE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, Accessed on September 20, 2022) (Higo et al. 1998). TBtools was used to generate heatmaps, and the R package gggenes was used to map the distribution of cis-acting elements (Chen et al. 2020; Gómez-Rubio 2017).
Gene duplication and evolutionary analysis
MCScanX-transposed was used to analyze the duplication type and calculate the synonymous substitution rate (Ks) and nonsynonymous substitution rate (Ka) (Wang et al. 2013). Subsequently, the differentiation time of the duplicated genes was further calculated according to T = Ks/2λ × 10–6 million years ago (Mya). λ is the substitution rate of 6.5 × 10–9 bases per synonymous substitution site per year (Li et al. 2023; Zhao et al. 2019a). Circos V0.69 was used to visualize gene duplication relationships (Krzywinski et al. 2009). MCScanX (python) was used to analyze the synteny of S. baicalensis with A. thaliana, Salvia miltiorrhiza, S. lycopersicum, O. sativa, Zea mays and Glycine max and for visualization (Wang et al. 2012).
RNA-seq analysis
Raw RNA-seq data for roots, stems, leaves, and flowers were downloaded from the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/view/PRJNA263255) (Hu et al. 2022). Sequences were requalified using fastp to filter out low-quality and linker sequences (Chen et al. 2018). The clean reads (75.26–81.99% of the total reads) were matched to the S. baicalensis CDSs using salmon, and the gene expression levels in the form of TPM (transcripts per kilobase of exon model per million mapped reads) were obtained (Patro et al. 2017). Subsequently, expression levels were displayed in the form of a heatmap by TBtools (Chen et al. 2020). Since this dataset had no biological replicates, for a more robust analysis of regulatory pathways, we downloaded the raw RNA-seq data of S. baicalensis roots, stems, and leaves published in earlier studies (https://www.ebi.ac.uk/ena/browser/text-search?query=PRJNA515574) (Gao et al. 2019). The TPM gene expression levels were obtained in the same way. The clean reads matched to CDSs accounted for 66.48–84.70% of the total reads.
Plants and treatments
S. baicalensis seeds of the same size with full grains were selected from Yangcheng County, Shanxi Province, China. The seeds were removed and placed in sterile water for imbibition for 12 h. Plants were grown under a 12 h photoperiod, day/night temperatures of 25/18 °C, an irradiance of 50 µmol m−2·s−1, and air humidity of 50%. Subsequent experiments were carried out after the seedlings had grown for 45 days.
S. baicalensis seedlings were used in abiotic stress and hormone treatment experiments according to Wang et al. (Wang et al. 2021). Details are as follows: (1) roots soaked in 20% (m/v) polyethylene glycol 6000 solution (PEG) to simulate drought; (2) 4 °C low temperature treatment; (3) roots soaked in 200 μM SA; (4) roots soaked in 100 μM ABA. Each treatment was replicated 3 times, with 10 seedlings per replicate. At 0, 3, 6, 12 and 24 h after treatment (HAT), 3 whole seedlings from the same replicate were randomly selected and mixed into one sample.
RT‒qPCR analysis of SbSPLs
The FastPure Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, China) was used to extract total RNA from entire S. baicalensis seedlings. Hifair® II 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) (Yeasen Biotechnology, Shanghai, China) was used for reverse transcription of the extracted RNA into cDNA. Hieff UNICON® qPCR SYBR Green Master Mix (Yeasen Biotechnology, Shanghai, China) was used for quantitative real-time polymerase chain reaction (RT‒qPCR). The S. baicalensis β-actin gene (GenBank: HQ847728) was used as the internal reference gene (Lu et al. 2022). Gene-specific primers for RT‒qPCR were independently designed based on nucleotide polymorphisms in the cDNA sequences of SbSPL1—SbSPL14 (Table S1). The RT‒qPCR procedure was as follows: predenaturation at 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 20 s; and detection of the signal at 72 °C. The 2−ΔΔCT method was used to calculate the expression levels. Three technical replicates were performed for each sample. Fisher’s least significant difference (LSD) method was used for difference analysis in Origin 2023 (Zhang et al. 2022b). The R package ggplot2 was used to draw histograms and annotate the significance analysis results (Gómez-Rubio 2017). The R package CluserGVis was used to conduct time series clustering analysis on the expression of SbSPLs under different treatments (Ni et al. 2020). By manually adjusting the number of clusters and observing the membership value, optimal clustering was achieved.
Prediction of S. baicalensis miR156/157 genes and their target SPL genes
To identify the miR156/157 genes in S. baicalensis, we downloaded the miR156/157 precursor sequences of A. thaliana from the miRBase database (Kozomara et al. 2019) and performed a homologous sequence search in the S. baicalensis genomic sequence using BLAST + (Shah et al. 2019). After manually removing redundant sequences, the secondary structure of the obtained sequences was predicted using MFold (http://www.mfold.org/, Accessed on 15 October 2022) (Zuker 2003). TBtools was used to map the chromosome location (Chen et al. 2020). In addition, we used the TAPIR tool (http://bioinformatics.psb.ugent.be/webtools/tapir/) to analyze SbSPLs potentially targeted by Sba-miR156/157 (Bonnet et al. 2010). The targeting relationship was displayed in the form of a heatmap by TBtools (Chen et al. 2020). Targeted SbSPLs were aligned with the reverse-complement sequences of Sba-miR156/157 using MAFFT (Nakamura et al. 2018). The miR156/157 sequences of A. thaliana and S. baicalensis were aligned in the same way. Alignment results were visualized using Texmaker (Beitz 2000).
Transcriptional regulation prediction and functional annotation
The 1.5 kb upstream sequence of the transcription start site of all genes was extracted, and the fimo tool of MEME Suite version 5.0.5 was used to predict the target genes that could bind to the conserved motifs of SbSPL proteins (Bailey et al. 2009). The coexpression coefficients of the SbSPLs and their possible target genes were calculated according to the expression levels. A correlation coefficient ≥ 0.5 (p value ≤ 0.05) represents a close positive correlation, and a correlation coefficient ≤ − 0.5 (p value ≤ 0.05) represents a close negative correlation. The CDS of S. baicalensis was uploaded to the Kyoto Encyclopedia of Genes and Genomes database (https://www.genome.jp/kegg/, Accessed on October 15, 2022) (Ogata et al. 1999), and A. thaliana was used as the reference species to obtain the functional annotation information of potential target genes. Cytoscape (v3.7.1) was used to visualize metabolic regulatory networks (Otasek et al. 2019).
Results
Chromosome distribution and basic protein information
We identified 14 SbSPLs and named them (SbSPL1—SbSPL14) according to chromosome positions (Figure S1). Except for Chr 3 and 5, SbSPLs were unevenly distributed on the other 7 chromosomes. SbSPLs were most distributed on Chr 2 and 8, with 3 each. Second, there are 2 SbSPLs on Chr 1, 7 and 9 and only 1 SbSPL on Chr 4 and 6. SbSPLs on Chr 1, 6, 7 and 8 were clearly localized to the distal telomeric regions of chromosomes.
The characteristics of SbSPL TFs are shown in Table S2. The protein sequence length varied greatly, ranging from 131 (SbSPL14) to 1076 (SbSPL8) amino acids (aa), and the MW ranged between 14.97 and 118.85 kDa. Most SbSPLs (SbSPL1/2/3/4/6/8/9/11/12/14) are basic (isoelectric point (Ip) > 7). The instability index (II) values were all greater than 40, and the grand average of hydropathicity (GRAVY) scores were all negative, indicating the instability and hydrophilicity of SbSPLs. Furthermore, proteins had different action sites. Subcellular localization predictions suggested that SbSPL2/3/5 were located outside the cell, while others were located in the nucleus (Table S2).
Phylogenetic and multiple sequence alignment analysis of the SbSPL TFs
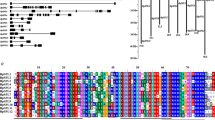
Fourteen SbSPL proteins were phylogenetically analyzed together with A. thaliana AtSPLs and divided into 8 groups (I–VIII) according to the grouping method of AtSPLs (Fig. 1). Groups III, IV, V and VII had the fewest members with one each, namely, SbSPL3, SbSPL5, SbSPL1 and SbSPL2. Group II had the most members (4), namely, SbSPL8/9/11/13. Groups I, VI and VIII each had 2 members, namely, SbSPL7/10, SbSPL12/14 and SbSPL4/6.
The integrity of the SBP domain (~ 76 aa) reflects the functionality of SbSPLs. As shown in Figure S2, the structural domain contained highly conserved sequences, such as CQQC, SCR, and RRR. Most SbSPLs contained 2 zinc finger domains (Zn-1 and Zn-2) and 1 NLS. Only Zn-1 of SbSPL2 (group VII) and Zn-2 of SbSPL4 (group VIII) were missing. Zn-1 of all Group I members (SbSPL7/10) was CCCC, while that of other members was CCCH. All SbSPLs contained a highly conserved NLS at the C-terminus of the SBP domain. The NLS of SbSPL3/7 was RRRR and that of other SbSPLs was RRRK.
Motif composition and gene structure
Motif and structure analysis further revealed the conservation of SbSPLs. We identified 10 conserved motifs (motif 1—motif 10) (Fig. 2B and Figure S3). Except for SbSPL2/4, which only had motif 1, the other members all contained motifs 1 and 2. Meanwhile, all members of Group I (SbSPL7/10) contained motifs 1, 2, 5 and 6; all members of group II (SbSPL8/9/11/13) contained all motifs. Similarly, members of Groups I and II contained the most introns (8–10) (Fig. 2C and Table S3). Groups III, IV, VI, VII and VIII members contain fewer introns (1–3). This suggested that genes with closer phylogenetic relationships have more similar structures.
Phylogenetic relationships, conserved motif composition and gene structure. A Phylogenetic tree. B Motif pattern of SbSPLs. The 10 colored boxes represent 10 different motifs, and their positions represent the positions on the protein. C Gene structure. CDS and introns are represented by green rectangles and black single lines, respectively
Analysis of cis-acting elements
Cis-acting element analysis was used to predict the potential biological functions of SbSPLs. The TATA box was the most numerous core promoter (801), followed by the CAAT box (584), and they were distributed in all SbSPL promoter regions (Fig. 3, Figure S4 and Table S4). Nine hormone-responsive cis-acting elements were identified. Most SbSPLs were related to the ABA pathway, and 8 SbSPL promoters contained 18 ABREs. In addition, 2 types of JA-responsive cis-acting elements (CGTCA motif and TGACG motif) were distributed in 8 SbSPL promoters, 14 of each type. The SA-responsive cis-acting element (TCA-element) was the least abundant, with only 4. Some stress-responsive cis-acting elements were also found in promoters. The promoters of SbSPL5/8/9/10/13 all had TC-rich repeats of defense and stress response elements. We also found 12 drought-induced elements (MBS), including 3 in SbSPL8/9, 2 in SbSPL10/13, and 1 in SbSPL12/14. Only SbSPL8/9/10 contained 1 low-temperature response element (LTR).
Replication, collinearity and Ka/Ks analysis
We analyzed the mechanisms of SbSPL expansion and evolution. Except for SbSPL2/11, the other SbSPLs participated in gene duplication events to form 8 gene pairs, including proximal duplication (1), segmental duplication (3) and transposable duplication (4) (Fig. 4). Segmental duplications and transposable duplications were the main causes of SbSPL expansion. The Ka/Ks values of all duplicate genes were less than 1 (0.3518–0.7839), indicating that SbSPLs experienced negative selection (Table S5). The divergence time of the duplicated SbSPLs ranged from 52.24 to 104.77 Mya.
Collinearity analysis of SbSPLs. The short lines of different colors inside the circle represent different gene duplication events, in which the short red lines represent proximal repeats; the short orange lines represent fragment repeats; the short green lines represent transposition repeats; and the short purple lines represent tandem repeats (none)
The syntenic gene pairs between species were analyzed. S. baicalensis had 13, 11, and 6 gene pairs with S. miltiorrhiza, S. lycopersicum, and A. thaliana, respectively (Fig. 5A–C). S. baicalensis had fewer syntenic gene pairs with the monocots Z. mays, S. bicolor, and O. sativa, with 1, 2, and 4 pairs, respectively (Fig. 5D–F). Orthologous genes of SbSPL4/6/10/14 were found in all dicots but not in monocots, indicating that these genes were formed after species divergence.
Tissue-specific expression analysis based on RNA-seq
We analyzed the expression pattern of SbSPLs in different tissues (root, stem, leaf, and flower) according to the published RNA-seq dataset (Fig. 6 and Table S6). Expression data for SbSPL4/5/7/11 was not obtained, suggesting that they may be pseudogenes or have specific spatiotemporal expression patterns not explored in this database. Among the 10 genes whose expression could be detected, SbSPL10 was most highly expressed in leaves and stems, while SbSPL1 and SbSPL13 were most highly expressed in roots and flowers, respectively. In addition, we also found that SbSPL9/10/12 were highly expressed in the 4 tissues.
Expression analysis of SbSPLs under abiotic stress and hormone treatment based on qPCR
To explore the potential functions of SbSPLs, the expression levels of 14 SbSPLs were analyzed at 5 time points (0, 3, 6, 12, and 24 HAT) under 4 °C, PEG, SA and ABA treatment. SbSPLs responded to 4 °C treatment earlier. The expression of 10 SbSPLs (SbSPL4/6/7/8/9/10/11/12/13/14) was upregulated at 3 HAT, downregulated at 6 and 12 HAT, and peaked at 24 h (Fig. 7A). In response to PEG, the expression of 11 SbSPLs (SbSPL2/3/4/5/7/8/9/10/12/13/14) began to significantly increase and peaked at 12 HAT (Fig. 7B). SbSPL7/9/10/12 were all found to respond extremely strongly to 4 °C and PEG. They increased by 4.18, 7.79, 4.62, and 5.79 times for 24 HAT at 4 °C and by 50.21, 61.48, 76.99, and 104.57 times for 12 HAT under PEG, respectively (Fig. 7A and B). This indicates the potential role of SbSPL7/9/10/12 in responding to a broad spectrum of abiotic stresses. Comparison of the fold differences also showed that the role of SbSPL7/9/10/12 under drought stress cannot be ignored. In addition, we also found that most SbSPLs responded to SA (14) and ABA (13) (Fig. 7C and D). Similar to 4 °C, 13 SbSPLs (SbSPL1–SbSPL13) were significantly upregulated and peaked at 24 HAT under ABA treatment, among which the expression levels of SbSPL3/6/7/9 were significantly increased by 8.77, 14, 9.5 and 53.44 times (Fig. 7D). In the SA treatment, the expression of SbSPL6/9 peaked at 3 HAT, increasing by 1.55 and 0.68 times, respectively; the expression of SbSPL4/8/11 peaked at 24 HAT, increasing by 0.72, 1.4 and 1.77 times, respectively (Fig. 7C). The expression levels of SbSPL1/3/4/5/7/10/12/13/14 were significantly downregulated at 3 HAT (Fig. 7C). Compared with SA, the response of SbSPLs to ABA was more regular and severe, which indicated the effectiveness of ABA in stimulating the function of SbSPLs.
Cluster analysis of time series was used to further search for similarity patterns in the expression of SbSPLs (Figure S5). In all treatments, when the number of clusters was 2, the membership value was higher than 0.5. Under abiotic stress (4 °C and PEG), SbSPL2/3/13, SbSPL5/6/10, and SbSPL4/7/8/9/11/12/14 showed consistent expression patterns (Figure S5 A and B). Under hormone-treated conditions (SA and ABA), the expression patterns of SbSPL2/8/11, SbSPL3/4/7/10/12/13, SbSPL5/14, and SbSPL6/9 were similar (Figure S5C and D). SbSPL3/13 and SbSPL4/12 were always similar in all treatments (Figure S5). This showed the functional consistency of SbSPLs.
Prediction of miR156/157 s regulating SbSPLs
We found 5 potential miR156s and 3 potential miR157s. They were distributed on chr 1 (Sba-miR157a/b), chr 2 (Sba-miR156a/b), chr 4 (Sba-miR156c/d), chr 5 (Sba-miR157c) and chr 9 (Sba-miR156e) and were named according to location (Figure S6). Sequence alignment analysis showed that the mature miR156/157 sequences of S. baicalensis were relatively conserved (Figure S7). The 8th base of the 3' end of miR156/157 s was U/A, and the 11th base contained U. Sba-miR157a/b/c inserted an A at the 11th base of the 5' end. In addition, the stems of the Sba-miR156/157 secondary structures all contained mature sequences (Figure S8). To analyze the posttranscriptional regulation pattern of SbSPLs, we predicted the SbSPLs targeted by Sba-miR156/157. Except for Sba-miR156b and Sba-miR157a, the other miR156/157 s may complementarily bind to the SbSPLs (Fig. 8 and Figure S9). SbSPL1–SbSPL5 had potential binding sites for Sba-miR156a/c/d/e, and miR156/157 complementary sites were relatively conserved among these SbSPLs (Fig. 8). Sequence differences were mainly limited to the 1/3/7 nucleotides of the complementary sequence.
Multiple sequence alignment of the reverse complementary sequences of Sba-miR156/157 and SbSPLs. Amino acid residues that are highly conserved in different sequences are marked with a blue background and an exclamation mark below; amino acid residues that are similar in different sequences are marked with a red background and an asterisk below. LOGO is above the aligned sequence
Prediction of SbSPLs on the regulation of phenylpropanoid and JA pathways
The 30,100 CDSs of S. baicalensis were submitted to the KEGG website, and A. thaliana was used as the reference species to determine metabolic pathways of S. baicalensis homologous genes. The MEME plugin fimo was used to predict target genes of SbSPLs, and potential downstream genes with similar expression patterns to SbSPLs were screened by correlation analysis (r ≥ 0.5&p value ≤ 0.05; r ≤ -0.5&p value ≤ 0.05). SbSPL-2/4/6 could bind to the upstream promoter of evm.model.contig106.199 (aeroate dehydratase, ADT), indicating that it may be involved in the synthesis of L-Phe (Fig. 9A and Table S7). The prediction results showed that the promoters of the JA synthesis pathway gene evm.model.contig321.52 (12-oxo-phytodienoic acid reductase, OPR), the phenylpropane metabolism pathway-related genes evm.model.contig515.180 (ferulate 5-hydroxylase, F5H) and evm.model.contig522.188 (caffeoyl-CoA oxymethyltransferase, CCoAOMT) were the binding targets of SbSPL2/6 (Fig. 9C and Table S7). In addition, POD homologous genes (evm.model.contig575.1, evm.model.contig300.287 and evm.model.contig507.297) had regulatory relationships with the rest of the genes except SbSPL13 (Fig. 9B and Table S7).
Regulatory network analysis. A Regulation of L-Phe biosynthesis by SbSPLs. B Regulation of lignin biosynthesis by SbSPLs. C Regulation of JA biosynthesis by SbSPLs. The red and blue lines represent positive and negative correlations, respectively. In the heatmap next to the gene name, red and blue represent higher and lower expression levels, respectively
Discussion
Characteristics and evolutionary mechanisms of SbSPLs
SPL TFs comprise a relatively small-scale family. Fourteen SbSPLs were identified from the S. baicalensis genome, and the number was similar to those of the dicotyledons A. thaliana (17) (Xu et al. 2016), Z. jujuba (18) (Shao et al. 2017), and F. vesca (14) (Xiong et al. 2018) and monocots O. sativa (19) (Xie et al. 2006) and S. italica (18) (Lai et al. 2022). The SPL sequences of S. baicalensis and the model species A. thaliana were highly conserved, and each group contained at least 1 AtSPL (Fig. 1). The sequence length (131–1076 aa) and MW (14.97 kDa -118.85 kDa) of SbSPL proteins varied greatly. This phenomenon was also observed in Vaccinium corymbosum VcSPLs (126–1072 aa; MW: 14.04–117.84 kDa) (Feng et al. 2023b), Prunus avium PavSPLs (162–1069 aa; MW: 18–118 kDa) (Sun et al. 2023) and Nicotiana tabacum NtSPLs (119–1001 aa; MW: 13.7–111.3 kDa) (He et al. 2023). SPL TFs diverged earlier than green algae (Guo et al. 2008) and initially formed two distinct lineages (clade I and clade II) in land plants. Clade I members had conserved structures, with more exons and longer protein sequences (Zhong et al. 2019). Groups I and II were consistent with clade 1, and the other groups could be placed in clade 2 (Fig. 2 and Table S2). Differentiation in genetic architecture partially explains the failure of SbSPL2/3/5 to localize to the nucleus.
The integrity and conservation of the SBP domain determine gene function (Xie et al. 2021). CCCH was mutated to CCCC in Zn-1 of SbSPL7/10 (Figure S2). The same changes occurred in A. thaliana AtSPL7 and S. italica SiSPL8 (Lai D et al. 2022) and CgoSPL6, DchSPL16, and GelSPL8 of three orchids (Zhao et al. 2023), indicating that Zn-1 could exist in 2 types (CCCC and CCCH). The Zn-1 of SbSPL2 and the Zn-2 sequence of SbSPL4 were missing (Figure S2). Zn-1 of FmSPL9 and Zn-2 of FmSPL31/32 were also absent in Fraxinus mandshurica (He et al. 2022a). Domain changes or losses are not always negative and drive the expansion of gene families (Brand et al. 2013; Wei et al. 2012).
Segmental duplication, tandem duplication and transposable duplication are considered to be the three principles of the evolutionary model (Moore RC and Purugganan 2003). Tandem duplication and segmental duplication are considered to be the main causes of gene family expansion (Liu et al. 2011). The expansion of SbSPLs mainly relied on transposable duplication (4) and segmental duplication (3) (Fig. 4 and Table S5). Duplications of Group I members (SbSPL7 and SbSPL10) and II (SbSPL8 and SbSPL9) had lower ka/ks (0.3350 and 0.3518) than other duplications (Table S5). Similar to previous results, the evolutionary speed of genes in Clade I seems to be slower (Zhu et al. 2020). The SbSPL repeat genes were subjected to purifying selection pressure (ka/ks < 1) (Table S5), ensuring the conservation of the protein. Transposable duplication genes had higher ka/ks (0.6387–0.7839) (Table S5). They faced higher selection pressure and were more evolutionarily active and were more likely to produce new functions or become pseudogenes (Wu et al. 2019). There was a strong positive correlation between species affinities and the synteny of SPLs. S. baicalensis had more orthologous SPLs with dicots than with monocots and had the most orthologous SPLs with the same family plant, S. bowleyana (13) (Fig. 5).
Potential functions of the SbSPLs
SPLs can regulate the development and growth of plant branches, flowers and other organs. Eight T. aestivum TaSPLs were found to be highly expressed in stems and inflorescences (Zhu et al. 2020). A. thaliana AtSPL2/9/11/13/15 promoted floral meristem recognition and floral induction (Xu et al. 2016). A. thaliana AtSPL1 and AtSPL12 were highly expressed in inflorescences, and their overexpression enhanced the heat tolerance of inflorescences (Chao et al. 2017) and belonged to group II with SbSPL13 (Fig. 1). SbSPL13 was extremely highly expressed in flowers, followed by SbSPL10, which was highly expressed in both flowers and stems (Fig. 6). The function of SbSPL13/10 in flower and stem development has research value.
Analysis of promoter cis-elements indicated that SbSPLs could be involved in abiotic stress and hormone responses (Fig. 3 and Table S4). SPL functions have been studied. Overexpression of OsSPL7 can improve the cold tolerance of O. sativa, while the knockout line showed strong low temperature sensitivity and growth inhibition (Hoang et al. 2019). AtSPL9 combined with the promoter of CBF2 to positively regulate cold resistance in A. thaliana, and miR156 balanced resistance responses and vegetative growth (Zhao et al. 2022). SbSPL4/6 belonged to group VIII with AtSPL9 (Fig. 1) and had a significant response to low temperature (Fig. 7A), indicating potential resistance. In addition, SbSPL4/6/7/8/9/10/11/12/13/14 showed a wave-like (increase–decrease-increase) expression pattern over time at low temperature (Fig. 7A), indicating that they could be regulated by factors such as miR156/157 to maintain the stability of growth and resistance responses. More notably, SbSPL7/9/10/12 responded strongly not only to low temperature but also to drought stress (Fig. 7B). Their identification provides candidate resistance genes to a broad spectrum of abiotic stresses. Several SPLs with drought resistance functions have been identified in multiple plants. MsSPL13 in Malus sieversii (Feng et al. 2023a), MsSPL8 in Medicago truncatula (Gou J et al. 2018), MeSPL9 in Manihot esculenta (Li et al. 2022b), and MsSPL9 in M. sativa (Hanly et al. 2020) negatively regulate drought resistance, while MsSPL13 in M. sativa (Arshad et al. 2017) and MiSPL13 in Mangifera indica (Zhu et al. 2022) enhance drought resistance. SbSPL1 is an orthologous gene of M. sieversii MsSPL13 (Feng et al. 2023a) and M. sativa MsSPL13 (Wang et al. 2019). However, the expression level of SbSPL1 did not change significantly under drought, indicating the species specificity of SPL functions. SbSPL2, SbSPL3 and SbSPL4/6 are orthologous genes of MiSPL13 (Zhu et al. 2022), MsSPL8 (Gou et al. 2018) and MeSPL9 (Li et al. 2022b), respectively. Their expression levels increased significantly at 12 HAT under drought, indicating potential function.
SA and ABA are important signaling molecules for plant physiological and morphological responses. They can regulate ROS levels and activate the expression of TFs and resistance genes during biotic and abiotic stress (Sah et al. 2016; Yang et al. 2023). SbSPLs differentially responded to SA; the expression levels of SbSPL1/3/4/5/7/10/12/13/14 were significantly downregulated at 3 HAT, while the expression levels of SbSPL6/9 and SbSPL4/8/11 were upregulated and reached a peak at 3 HAT and 24 HAT, respectively (Fig. 7C). This suggested their differentiated functions. The ABA pathway is the core of plant drought resistance, and ABREs are conserved in the promoters of drought-responsive genes (Muhammad Aslam et al. 2022; Waadt et al. 2022). The promoters of 8 SbSPLs contained ABREs (18) (Fig. 3, Figure S4 and Table S4), and 13 SbSPLs were upregulated in response to ABA (Fig. 7D), indicating possible drought resistance. Most SbSPLs responded to drought earlier than to ABA (Fig. 7A and Fig. 7D, Figure S5). It is speculated that there is a complex regulatory mechanism between drought, ABA and SbSPLs. Furthermore, the strong response of SbSPL9 to stress and ABA suggests its importance (Fig. 7). In summary, our study showed that SbSPLs respond to abiotic stress and hormones, which provides a reference for subsequent research on gene functions.
The potential regulation of SbSPLs by miR156/157 and its potential impact on the accumulation of downstream substances
SPLs in plants often coordinate with miR156/157 to function together. Eleven of the 17 AtSPLs were miR156 targets in A. thaliana (Xu et al. 2016). Eleven OsSPL genes were predicted targets of OsmiR156 in O. sativa (Xie et al. 2006). Similar to previous studies, SbSPL1—SbSPL5 were potential target genes of Sba-miR156/157 (Fig. 8 and Figure S9).
SPL TFs serve as activators or inhibitors of gene expression and are involved in regulating the synthesis of plant metabolites (Kajla et al. 2023). Morus alba MnSPL7 increased the expression levels of catechin synthesis genes (f3’h, DFR and LAR) by promoting the transcription of MnTT2L2 (Li et al. 2022a). Artemisia annua AaSPL2 and DBR2 act synergistically at the “GTAC” cis-element in the DBR2 promoter to mediate transcriptional activation of DBR2 in response to JA, thereby increasing artemisinin content (Lv et al. 2019). In this study, SbSPL2/4/6 was closely related to the expression of evm.model.contig106.199 (ADT), a key gene for L-Phe synthesis (Fig. 9A and Table S7). L-Phe is the starting material in the synthesis pathway of phenylpropanoids and flavonoids, which is bound to affect flavonoid accumulation (Dong and Lin 2021). In addition, lignin is important for plant environmental adaptability. Several recent studies have reported lignin to be involved in plant responses to stress. MeRAV5 activates cassava lignin accumulation and improves drought resistance (Yan et al. 2021), and knocking out the F5H gene to reduce S-lignin/G-lignin improved S. baicalensis resistance to Brassica napus and increased stem strength (Cao et al. 2022). Our results predicted that the F5H (evm.model.contig515.180) and CCoAOMT (evm.model.contig522.188) genes were positively regulated by SbSPL2/6. The opposite state appeared in the regulation of F5H (evm.model.contig515.180) by SbSPL3 (Fig. 9B and Table S7). We speculate that SbSPLs regulate key genes of lignin synthesis and affect the ratio of G-ligin and S-ligin, thereby affecting the stress resistance of S. baicalensis. Notably, PODs (evm.model.contig575.1, evm.model.contig300.287 and evm.model.contig507.297) appeared to be more complexly regulated by SbSPLs (Fig. 9B and Table S7). As a key enzyme system in the oxidative stress response, POD has unique functions in controlling various aspects of plant growth, development, cell metabolism and defense signal transduction (Lee et al. 2018). Another important pathway in the defense response is the synthesis and signal transduction of JA. Key gene expression of SPL7-regulated JA synthesis in A. thaliana has been demonstrated (Yan et al. 2017). Likewise, the miR156-SPL9 module positively regulated A. thaliana resistance to Botrytis cinerea through the JA pathway (Sun et al. 2022). In our study, the binding of SbSPL2/4/6/11 to the OPR (evm.model.contig321.52) promoter (Fig. 9C, Table S7) and similar expression patterns might provide another case.
Conclusion
In this study, 14 SbSPLs with SBP-box domains were identified and divided into 8 groups based on phylogenetic analysis. The genetic structures of SbSPLs in the same group were highly similar. Five SbSPLs contained complementary sequences of miR156/157 s and could be used as potential targets. In addition, SbSPLs contained abiotic stress- and hormone-responsive cis-acting elements. Gene expression pattern analysis based on transcriptional profiling and qPCR demonstrated that SbSPLs potentially regulated the development and growth of organs, and they (especially SbSPL9) responded to low temperature, drought, SA, and ABA. These results increase the understanding of the evolution and biological importance of SbSPLs and provide a reference for revealing the functional characteristics of SbSPLs and the mechanism for genetic improvement of plants under stress.
Data availability
All data generated or analyzed in this study are included in this published article and its Supplementary Material. The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request. The analysis websites used in this study are as follows: The DNA and protein sequence information of S. baicalensis was downloaded from the National Genomics Data Center (https://ngdc.cncb.ac.cn/, accession number: PRJCA009554). Raw RNA-Seq data for roots, stems, leaves, and flowers were downloaded from the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/home, accession number: PRJNA263255). All clean sequence read data of S. baicalensis roots, stems, and leaves were deposited in the NCBI SRA database (https://www.ncbi.nlm.nih.gov/sra, accession number: PRJNA515574). Pfam database (http://pfam.sanger.ac.uk/), SMART (http://smart.embl.de/), ExPASy Bioinformatics Resource Portal (https://web.expasy.org/compute_pi/), ProtComp 9.0 (http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc), PLACE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/), MFold (http://www.mfold.org/), Kyoto Encyclopedia of Genes and Genomes database (https://www.genome.jp/kegg/).
References
Arshad M, Feyissa BA, Amyot L, Aung B, Hannoufa A (2017) MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci 258:122–136. https://doi.org/10.1016/j.plantsci.2017.01.018
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Beitz E (2000) TEXshade: shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics 16:135–139. https://doi.org/10.1093/bioinformatics/16.2.135
Birkenbihl RP, Jach G, Saedler H, Huijser P (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352:585–596. https://doi.org/10.1016/j.jmb.2005.07.013
Bonnet E, He Y, Billiau K, Van de Peer Y (2010) TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26:1566–1568. https://doi.org/10.1093/bioinformatics/btq233
Brand LH, Fischer NM, Harter K, Kohlbacher O, Wanke D (2013) Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res 41:9764–9778. https://doi.org/10.1093/nar/gkt732
Cao Y, Yan X, Ran S, Ralph J, Smith RA, Chen X, Qu C, Li J, Liu L (2022) Knockout of the lignin pathway gene BnF5H decreases the S/G lignin compositional ratio and improves Sclerotinia sclerotiorum resistance in Brassica napus. Plant Cell Environ 45:248–261. https://doi.org/10.1111/pce.14208
Cardon G, Höhmann S, Klein J, Nettesheim K, Saedler H, Huijser P (1999) Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237:91–104. https://doi.org/10.1016/s0378-1119(99)00308-x
Chao LM, Liu YQ, Chen DY, Xue XY, Mao YB, Chen XY (2017) Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol Plant 10:735–748. https://doi.org/10.1016/j.molp.2017.03.010
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Consortium TU (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158-d169. https://doi.org/10.1093/nar/gkw1099
Dong NQ, Lin HX (2021) Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J Integr Plant Biol 63:180–209. https://doi.org/10.1111/jipb.13054
Duvaud S, Gabella C, Lisacek F, Stockinger H, Ioannidis V, Durinx C (2021) Expasy, the swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res 49:W216-w227. https://doi.org/10.1093/nar/gkab225
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016. https://doi.org/10.1006/jmbi.2000.3903
Feng G, Han J, Yang Z, Liu Q, Shuai Y, Xu X, Nie G, Huang L, Liu W, Zhang X (2021) Genome-wide identification, phylogenetic analysis, and expression analysis of the SPL gene family in orchardgrass (Dactylis glomerata L.). Genomics 113:2413–2425. https://doi.org/10.1016/j.ygeno.2021.05.032
Feng C, Zhang X, Du B, Xiao Y, Wang Y, Sun Y, Zhou X, Wang C, Liu Y, Li TH (2023a) MicroRNA156ab regulates apple plant growth and drought tolerance by targeting transcription factor MsSPL13. Plant Physiol 192:1836–1857. https://doi.org/10.1093/plphys/kiad099
Feng X, Zhou B, Wu X, Wu H, Zhang S, Jiang Y, Wang Y, Zhang Y, Cao M, Guo B, Su S, Hou Z (2023b) Molecular characterization of SPL gene family during flower morphogenesis and regulation in blueberry. BMC Plant Biol 23:40. https://doi.org/10.1186/s12870-023-04044-x
Gao T, Xu Z, Song X, Huang K, Li Y, Wei J, Zhu X, Ren H, Sun C (2019) Hybrid sequencing of full-length cDNA transcripts of the medicinal plant Scutellaria baicalensis. Int J Mol Sci. https://doi.org/10.3390/ijms20184426
Gómez-Rubio V (2017) ggplot2 - Elegant graphics for data analysis (2nd Edition). J Stat Softw 77:1–3. https://doi.org/10.18637/jss.v077.b02
Gou J, Debnath S, Sun L, Flanagan A, Tang Y, Jiang Q, Wen J, Wang ZY (2018) From model to crop: functional characterization of SPL8 in M. truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in alfalfa. Plant Biotechnol J 16:951–962. https://doi.org/10.1111/pbi.12841
Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J (2008) Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418:1–8. https://doi.org/10.1016/j.gene.2008.03.016
Guo L, Wang S, Zhang J, Yang G, Zhao M, Ma W, Zhang X, Li X, Han B, Chen N, Huang L (2013) Effects of ecological factors on secondary metabolites and inorganic elements of Scutellaria baicalensis and analysis of geoherblism. Sci China Life Sci 56:1047–1056. https://doi.org/10.1007/s11427-013-4562-5
Hanly A, Karagiannis J, Lu QSM, Tian L, Hannoufa A (2020) Characterization of the role of SPL9 in drought stress tolerance in Medicago sativa. Int J Mol Sci. https://doi.org/10.3390/ijms21176003
He B, Gao S, Lu H, Yan J, Li C, Ma M, Wang X, Chen X, Zhan Y, Zeng F (2022a) Genome-wide analysis and molecular dissection of the SPL gene family in Fraxinus mandshurica. BMC Plant Biol 22:451. https://doi.org/10.1186/s12870-022-03838-9
He F, Long R, Wei C, Zhang Y, Li M, Kang J, Yang Q, Wang Z, Chen L (2022b) Genome-wide identification, phylogeny and expression analysis of the SPL gene family and its important role in salt stress in Medicago sativa L. BMC Plant Biol 22:295. https://doi.org/10.1186/s12870-022-03678-7
He L, Peng X, Cao H, Yang K, Xiang L, Li R, Zhang F, Liu W (2023) The NtSPL gene family in Nicotiana tabacum: genome-wide investigation and expression analysis in response to cadmium stress. Genes. https://doi.org/10.3390/genes14010183
Higo K, Ugawa Y, Iwamoto M, Higo H (1998) PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res 26:358–359. https://doi.org/10.1093/nar/26.1.358
Hoang TV, Vo KTX, Rahman MM, Choi SH, Jeon JS (2019) Heat stress transcription factor OsSPL7 plays a critical role in reactive oxygen species balance and stress responses in rice. Plant Sci 289:110273. https://doi.org/10.1016/j.plantsci.2019.110273
Hou H, Jia H, Yan Q, Wang X (2018) Overexpression of a SBP-Box gene (VpSBP16) from Chinese wild vitis species in Arabidopsis improves salinity and drought stress tolerance. Int J Mol Sci. https://doi.org/10.3390/ijms19040940
Hu S, Wang D, Wang W, Zhang C, Li Y, Wang Y, Zhou W, Niu J, Wang S, Qiang Y, Cao X, Wang Z (2022) Whole genome and transcriptome reveal flavone accumulation in Scutellaria baicalensis roots. Front Plant Sci 13:1000469. https://doi.org/10.3389/fpls.2022.1000469
Huang X, Wu P, Huang F, Xu M, Chen M, Huang K, Li GP, Xu M, Yao D, Wang L (2017) Baicalin attenuates chronic hypoxia-induced pulmonary hypertension via adenosine A(2A) receptor-induced SDF-1/CXCR4/PI3K/AKT signaling. J Biomed Sci 24:52. https://doi.org/10.1186/s12929-017-0359-3
Kajla M, Roy A, Singh IK, Singh A (2023) Regulation of the regulators: transcription factors controlling biosynthesis of plant secondary metabolites during biotic stresses and their regulation by miRNAs. Front Plant Sci 14:1126567. https://doi.org/10.3389/fpls.2023.1126567
Klein J, Saedler H, Huijser P (1996) A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet 250:7–16. https://doi.org/10.1007/bf02191820
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155-d162. https://doi.org/10.1093/nar/gky1141
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. https://doi.org/10.1101/gr.092759.109
Lai D, Fan Y, Xue G, He A, Yang H, He C, Li Y, Ruan J, Yan J, Cheng J (2022) Genome-wide identification and characterization of the SPL gene family and its expression in the various developmental stages and stress conditions in foxtail millet (Setaria italica). BMC Genomics 23:389. https://doi.org/10.1186/s12864-022-08633-2
Lee ES, Kang CH, Park JH, Lee SY (2018) Physiological significance of plant peroxiredoxins and the structure-related and multifunctional biochemistry of peroxiredoxin 1. Antioxid Redox Signal 28:625–639. https://doi.org/10.1089/ars.2017.7400
Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493-d496. https://doi.org/10.1093/nar/gkx922
Li H, Ma B, Luo Y, Wei W, Yuan J, Zhai C, He N (2022a) The Mulberry SPL Gene Family and the Response of MnSPL7 to Silkworm Herbivory through Activating the Transcription of MnTT2L2 in the Catechin Biosynthesis Pathway. Int J Mol Sci. https://doi.org/10.3390/ijms23031141
Li S, Cheng Z, Li Z, Dong S, Yu X, Zhao P, Liao W, Yu X, Peng M (2022b) MeSPL9 attenuates drought resistance by regulating JA signaling and protectant metabolite contents in cassava. Theor Appl Genet 135:817–832. https://doi.org/10.1007/s00122-021-04000-z
Li H, Wu S, Lin R, Xiao Y, Malaco Morotti AL, Wang Y, Galilee M, Qin H, Huang T, Zhao Y, Zhou X, Yang J, Zhao Q, Kanellis AK, Martin C, Tatsis EC (2023) The genomes of medicinal skullcaps reveal the polyphyletic origins of clerodane diterpene biosynthesis in the family Lamiaceae. Mol Plant 16:549–570. https://doi.org/10.1016/j.molp.2023.01.006
Liu H, Yang W, Liu D, Han Y, Zhang A, Li S (2011) Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol Biol Rep 38:417–427. https://doi.org/10.1007/s11033-010-0124-0
Liu H, Ye F, Sun Q, Liang H, Li C, Li S, Lu R, Huang B, Tan W, Lai L (2021) Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J Enzyme Inhib Med Chem 36:497–503. https://doi.org/10.1080/14756366.2021.1873977
Lopez-Ortiz C, Peña-Garcia Y, Bhandari M, Abburi VL, Natarajan P, Stommel J, Nimmakayala P, Reddy UK (2021) Identification of miRNAs and their targets involved in flower and fruit development across domesticated and wild Capsicum species. Int J Mol Sci. https://doi.org/10.3390/ijms22094866
Lu Y, Cao B, Su Y, Yang J, Xue Y, Zhang M, Che L, Gao P, Li X, Zhou X, Liu L, Song S, Li G, Bai C (2022) Inter-specific differences of medicinal bioactive products are correlated with differential expressions of key enzyme genes in Scutellaria baicalensis and Scutellaria viscidula. Ind Crops Prod 189:115758. https://doi.org/10.1016/j.indcrop.2022.115758
Lv Z, Wang Y, Liu Y, Peng B, Zhang L, Tang K, Chen W (2019) The SPB-Box transcription factor AaSPL2 positively regulates artemisinin biosynthesis in Artemisia annua L. Front Plant Sci 10:409. https://doi.org/10.3389/fpls.2019.00409
Malekmohammad K, Rafieian-Kopaei M (2021) Mechanistic aspects of medicinal plants and secondary metabolites against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Curr Pharm Des 27:3996–4007. https://doi.org/10.2174/1381612827666210705160130
Matthews C, Arshad M, Hannoufa A (2019) Alfalfa response to heat stress is modulated by microRNA156. Physiol Plant 165:830–842. https://doi.org/10.1111/ppl.12787
Meng C, Wang P, Hao Z, Gao Z, Li Q, Gao H, Liu Y, Li Q, Wang Q, Feng F (2022) Ecological and health risk assessment of heavy metals in soil and Chinese herbal medicines. Environ Geochem Health 44:817–828. https://doi.org/10.1007/s10653-021-00978-z
Moore RC, Purugganan MD (2003) The early stages of duplicate gene evolution. PNAS 100:15682–15687. https://doi.org/10.1073/pnas.2535513100
Muhammad Aslam M, Waseem M, Jakada BH, Okal EJ, Lei Z, Saqib HSA, Yuan W, Xu W, Zhang Q (2022) Mechanisms of abscisic acid-mediated drought stress responses in plants. Int J Mol Sci. https://doi.org/10.3390/ijms23031084
Na HY, Lee BC (2019) Scutellaria baicalensis alleviates insulin resistance in diet-induced obese mice by modulating inflammation. Int J Mol Sci. https://doi.org/10.3390/ijms20030727
Nakamura T, Yamada KD, Tomii K, Katoh K (2018) Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34:2490–2492. https://doi.org/10.1093/bioinformatics/bty121
Ni J, Zhao Y, Tao R, Yin L, Gao L, Strid Å, Qian M, Li J, Li Y, Shen J, Teng Y, Bai S (2020) Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol J 18:1223–1240. https://doi.org/10.1111/pbi.13287
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27:29–34. https://doi.org/10.1093/nar/27.1.29
Otasek D, Morris JH, Bouças J, Pico AR, Demchak B (2019) Cytoscape Automation: empowering workflow-based network analysis. Genome Biol 20:185. https://doi.org/10.1186/s13059-019-1758-4
Park HJ, Park SH, Choi YH, Chi GY (2021) The root extract of Scutellaria baicalensis induces apoptosis in EGFR TKI-resistant human lung cancer cells by inactivation of STAT3. Int J Mol Sci. https://doi.org/10.3390/ijms22105181
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. https://doi.org/10.1038/nmeth.4197
Perea-García A, Andrés-Bordería A, Huijser P, Peñarrubia L (2021) The copper-microRNA pathway Is integrated with developmental and environmental stress responses in Arabidopsis thaliana. Int J Mol Sci. https://doi.org/10.3390/ijms22179547
Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD (2018) HMMER web server: 2018 update. Nucleic Acids Res 46:W200-w204. https://doi.org/10.1093/nar/gky448
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. https://doi.org/10.1093/molbev/msp077
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD (2012) The Pfam protein families database. Nucleic Acids Res 40:D290-301. https://doi.org/10.1093/nar/gkr1065
Quinlan AR (2014) BEDTools: the swiss-army tool for genome feature analysis. Curr Protoc Bioinform 47:11.12.1-34. https://doi.org/10.1002/0471250953.bi1112s47
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520. https://doi.org/10.1016/s0092-8674(02)00863-2
Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7:571. https://doi.org/10.3389/fpls.2016.00571
Salinas M, Xing S, Höhmann S, Berndtgen R, Huijser P (2012) Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 235:1171–1184. https://doi.org/10.1007/s00425-011-1565-y
Shah N, Nute MG, Warnow T, Pop M (2019) Misunderstood parameter of NCBI BLAST impacts the correctness of bioinformatics workflows. Bioinformatics 35:1613–1614. https://doi.org/10.1093/bioinformatics/bty833
Shao F, Lu Q, Wilson IW, Qiu D (2017) Genome-wide identification and characterization of the SPL gene family in Ziziphus jujuba. Gene 627:315–321. https://doi.org/10.1016/j.gene.2017.06.044
Song JW, Long JY, Xie L, Zhang LL, Xie QX, Chen HJ, Deng M, Li XF (2020) Applications, phytochemistry, pharmacological effects, pharmacokinetics, toxicity of Scutellaria baicalensis Georgi. and its probably potential therapeutic effects on COVID-19: a review. Chin Med 15:102. https://doi.org/10.1186/s13020-020-00384-0
Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26:1792–1807. https://doi.org/10.1105/tpc.114.123851
Su HX, Yao S, Zhao WF, Li MJ, Liu J, Shang WJ, Xie H, Ke CQ, Hu HC, Gao MN, Yu KQ, Liu H, Shen JS, Tang W, Zhang LK, Xiao GF, Ni L, Wang DW, Zuo JP, Jiang HL, Bai F, Wu Y, Ye Y, Xu YC (2020) Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol Sin 41:1167–1177. https://doi.org/10.1038/s41401-020-0483-6
Sun T, Zhou Q, Zhou Z, Song Y, Li Y, Wang HB, Liu B (2022) SQUINT positively regulates resistance to the pathogen Botrytis cinerea via miR156-SPL9 module in Arabidopsis. Plant Cell Physiol 63:1414–1432. https://doi.org/10.1093/pcp/pcac042
Sun Y, Wang Y, Xiao Y, Zhang X, Du B, Turupu M, Wang C, Yao Q, Gai S, Huang J, Tong S, Li T (2023) Genome-wide identification of the SQUAMOSA promoter-binding protein-like (SPL) transcription factor family in sweet cherry fruit. Int J Mol Sci. https://doi.org/10.3390/ijms24032880
Waadt R, Seller CA, Hsu PK, Takahashi Y, Munemasa S, Schroeder JI (2022) Plant hormone regulation of abiotic stress responses. Nat Rev Mol Cell Biol 23:680–694. https://doi.org/10.1038/s41580-022-00479-6
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49. https://doi.org/10.1093/nar/gkr1293
Wang Y, Li J, Paterson AH (2013) MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 29:1458–1460. https://doi.org/10.1093/bioinformatics/btt150
Wang H, Lu Z, Xu Y, Kong L, Shi J, Liu Y, Fu C, Wang X, Wang ZY, Zhou C, Han L (2019) Genome-wide characterization of SPL family in Medicago truncatula reveals the novel roles of miR156/SPL module in spiky pod development. BMC Genomics 20:552. https://doi.org/10.1186/s12864-019-5937-1
Wang W, Hu S, Cao Y, Chen R, Wang Z, Cao X (2021) Selection and evaluation of reference genes for qRT-PCR of Scutellaria baicalensis Georgi under different experimental conditions. Mol Biol Rep 48:1115–1126. https://doi.org/10.1007/s11033-021-06153-y
Wang W, Hu S, Zhang C, Yang J, Zhang T, Wang D, Cao X, Wang Z (2022) Systematic analysis and functional characterization of R2R3-MYB genes in Scutellaria baicalensis Georgi. Int J Mol Sci. https://doi.org/10.3390/ijms23169342
Wani SH, Anand S, Singh B, Bohra A, Joshi R (2021) WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep 40:1071–1085. https://doi.org/10.1007/s00299-021-02691-8
Wei KF, Chen J, Chen YF, Wu LJ, Xie DX (2012) Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res 19:153–164. https://doi.org/10.1093/dnares/dsr048
Wen S, Zhang X, Wu Y, Yu S, Zhang W, Liu D, Yang K, Sun J (2022) Agrimonia pilosa Ledeb.: a review of its traditional uses, botany, phytochemistry, pharmacology, and toxicology. Heliyon 8:e09972. https://doi.org/10.1016/j.heliyon.2022.e09972
Wu D, Luo J, Chen J, Zhang L, Lim KJ, Wang Z (2019) Selection pressure causes differentiation of the SPL gene family in the Juglandaceae. Mol Genet Genomics 294:1037–1048. https://doi.org/10.1007/s00438-019-01562-y
Xiang L, Gao Y, Chen S, Sun J, Wu J, Meng X (2022) Therapeutic potential of Scutellaria baicalensis Georgi in lung cancer therapy. Phytomedicine 95:153727. https://doi.org/10.1016/j.phymed.2021.153727
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142:280–293. https://doi.org/10.1104/pp.106.084475
Xie X, Yue S, Shi B, Li H, Cui Y, Wang J, Yang P, Li S, Li X, Bian S (2021) Comprehensive analysis of the SBP family in blueberry and their regulatory mechanism controlling chlorophyll accumulation. Front Plant Sci 12:703994. https://doi.org/10.3389/fpls.2021.703994
Xiong JS, Zheng D, Zhu HY, Chen JQ, Na R, Cheng ZM (2018) Genome-wide identification and expression analysis of the SPL gene family in woodland strawberry Fragaria vesca. Genome 61:675–683. https://doi.org/10.1139/gen-2018-0014
Xu M, Hu T, Zhao J, Park MY, Earley KW, Wu G, Yang L, Poethig RS (2016) Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12:e1006263. https://doi.org/10.1371/journal.pgen.1006263
Yan J, Chia J-C, Sheng H, Jung H-i, Zavodna T-O, Zhang L, Huang R, Jiao C, Craft EJ, Fei Z, Kochian LV, Vatamaniuk OK (2017) Arabidopsis pollen fertility requires the transcription factors CITF1 and SPL7 that regulate copper delivery to anthers and jasmonic acid synthesis. Plant Cell 29:3012–3029. https://doi.org/10.1105/tpc.17.00363
Yan Y, Wang P, Lu Y, Bai Y, Wei Y, Liu G, Shi H (2021) MeRAV5 promotes drought stress resistance in cassava by modulating hydrogen peroxide and lignin accumulation. Plant J 107:847–860. https://doi.org/10.1111/tpj.15350
Yang H, Fang R, Luo L, Yang W, Huang Q, Yang C, Hui W, Gong W, Wang J (2023) Uncovering the mechanisms of salicylic acid-mediated abiotic stress tolerance in horticultural crops. Front Plant Sci. https://doi.org/10.3389/fpls.2023.1226041
Yu G, Lam TT-Y, Zhu H, Guan Y (2018) Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol 35:3041–3043. https://doi.org/10.1093/molbev/msy194
Yuan Y, Shuai L, Chen S, Huang L, Qin S, Yang Z (2012) Flavonoids and antioxidative enzymes in temperature-challenged roots of Scutellaria baicalensis Georgi. Z Naturforsch C J Biosci 67:77–85. https://doi.org/10.1515/znc-2012-1-210
Yun C, Zhao Z, Ri I, Gao Y, Shi Y, Miao N, Gu L, Wang W, Wang H (2022) How does UV-B stress affect secondary metabolites of Scutellaria baicalensis in vitro shoots grown at different 6-benzyl aminopurine concentrations? Physiol Plant 174:e13778. https://doi.org/10.1111/ppl.13778
Zhang YG, Han M, Jiang X, Zhao SN, Yang LM (2013) Physiological ecology responses of Scutellaria baicalensis to drought and rewatering. Zhongguo Zhong Yao Za Zhi 38:3845–3850
Zhang C, Wang W, Wang D, Hu S, Zhang Q, Wang Z, Cui L (2022a) Genome-wide identification and characterization of the WRKY Gene Family in Scutellaria baicalensis Georgi under diverse abiotic stress. Int J Mol Sci. https://doi.org/10.3390/ijms23084225
Zhang F, Li C, Qu X, Liu J, Yu Z, Wang J, Zhu J, Yu Y, Ding Z (2022b) A feedback regulation between ARF7-mediated auxin signaling and auxin homeostasis involving MES17 affects plant gravitropism. J Integr Plant Biol 64:1339–1351. https://doi.org/10.1111/jipb.13268
Zhao Q, Chen X-Y, Martin C (2016) Scutellaria baicalensis, the golden herb from the garden of Chinese medicinal plants. Sci Bull 61:1391–1398. https://doi.org/10.1007/s11434-016-1136-5
Zhao Q, Yang J, Cui MY, Liu J, Fang Y, Yan M, Qiu W, Shang H, Xu Z, Yidiresi R, Weng JK, Pluskal T, Vigouroux M, Steuernagel B, Wei Y, Yang L, Hu Y, Chen XY, Martin C (2019a) The reference genome sequence of Scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis. Mol Plant 12:935–950. https://doi.org/10.1016/j.molp.2019.04.002
Zhao T, Tang H, Xie L, Zheng Y, Ma Z, Sun Q, Li X (2019b) Scutellaria baicalensis Georgi. (Lamiaceae): a review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. J Pharm Pharmacol 71:1353–1369. https://doi.org/10.1111/jphp.13129
Zhao J, Shi M, Yu J, Guo C (2022) SPL9 mediates freezing tolerance by directly regulating the expression of CBF2 in Arabidopsis thaliana. BMC Plant Biol 22:59. https://doi.org/10.1186/s12870-022-03445-8
Zhao X, Zhang M, He X, Zheng Q, Huang Y, Li Y, Ahmad S, Liu D, Lan S, Liu Z (2023) Genome-wide identification and expression analysis of the SPL gene family in three orchids. Int J Mol Sci. https://doi.org/10.3390/ijms241210039
Zhong H, Kong W, Gong Z, Fang X, Deng X, Liu C, Li Y (2019) Evolutionary analyses reveal diverged patterns of SQUAMOSA promoter binding protein-like (SPL) gene family in oryza genus. Front Plant Sci 10:565. https://doi.org/10.3389/fpls.2019.00565
Zhou Q, Shi J, Li Z, Zhang S, Zhang S, Zhang J, Bao M, Liu G (2021) miR156/157 targets SPLs to regulate flowering transition, plant architecture and flower organ size in petunia. Plant Cell Physiol 62:839–857. https://doi.org/10.1093/pcp/pcab041
Zhu T, Liu Y, Ma L, Wang X, Zhang D, Han Y, Ding Q, Ma L (2020) Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol 20:420. https://doi.org/10.1186/s12870-020-02576-0
Zhu J-w, He X-h, Li Y-z, Zhang Y-l, Yu H-x, Xia L-m, Mo X, Zeng X-m, Yang J-h, Luo C (2022) Genome-wide analysis of the mango SPL family and overexpression of MiSPL13 confers early flowering and stress tolerance in transgenic Arabidopsis. Sci Hortic 305:111363. https://doi.org/10.1016/j.scienta.2022.111363
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. https://doi.org/10.1093/nar/gkg595
Acknowledgements
Thanks to all co-authors for their dedication to this article. The authors thank the editors and reviewers for their work in promoting the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32000264) and Guangxi Key Laboratory of Traditional Chinese Medicine Quality Standards Systematic research topics (GZZX201805) and Survey and Collection of Germplasm Resources of Woody Herbaceous Plants in Guangxi, China (GXFS-2021-34).
Author information
Authors and Affiliations
Contributions
Conceptualization, Z.Z. and H.Y.; methodology, W.J. and H.Y.; software, W.J. and Z.Z.; validation, Z.Z., W.J. and H.R.; formal analysis, H.R.; investigation, H.R.; resources, Z.Z.; data curation, Z.Z. and W.J.; writing—original draft preparation, Z.Z.; writing—review and editing, W.J., H.R. and H.Y.; visualization, Z.Z and W.J.; supervision, H.R. and H.Y.; project administration, H.R. and H.Y.; funding acquisition, H.R. and H.Y.; manuscript structure revision for major revisions, W.J.; manuscript content reduction for major revisions, W.J.; manuscript error correction for major revisions, W.J.; manuscript format modification for major revisions, W.J.; point-by-point responses to reviewer questions for major revisions, W.J.; preliminary polishing of the manuscript and contacting the company to polish the manuscript, W.J.; inspection of all materials for major revisions, W.J., Z.Z., H.R. and H.Y. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Ethical approval
Plants materials involved in this research are used for scientific research and are allowed to be used and provided free of charge in this study. This article did not contain any studies with human participants or animals and did not involve any endangered or protected species.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, Jw., Zhao, Zy., Hu, Rc. et al. Genome-wide identification, stress- and hormone-responsive expression characteristics, and regulatory pattern analysis of Scutellaria baicalensis SbSPLs. Plant Mol Biol 114, 20 (2024). https://doi.org/10.1007/s11103-023-01410-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-023-01410-z