Abstract
Purpose
To evaluate a random forest model that counts silicone oil droplets and non-silicone oil particles in protein formulations with large class imbalance.
Methods
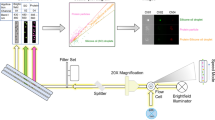
In this work, we present a novel approach for automated image analysis of flow microscopy data based on random forest classification enabling rapid analysis of large data sets. The random forest approach overcomes many of the limitations of traditional classification schemes derived from simple filters or regression models. In particular, the approach does not require a priori selection of important morphology parameters.
Results
We analyzed silicone oil droplets and non-silicone oil particles observed in four model systems with protein concentrations of 20, 50 and 125 mg/mL. Filters based on random forests achieve higher classification accuracies when compared to regression based filters. Additionally, we showcase a procedure that allows for accurate counting of particles ≥1 μm.
Conclusions
Our method is generally applicable for classification and counting of different classes of particles as long as class morphologies are differentially expressed.







Similar content being viewed by others
Notes
The density histogram is a smoothed version of the relative histogram such that the entire area of the histogram equals 1.
Abbreviations
- CART:
-
Classification and regression tree
- ECD:
-
Equivalent cirlcular diameter
- ESD:
-
Equivalent spherical diameter
- mAb:
-
Monoclonal antibody
- NSO:
-
Non-silicone oil
- PFS:
-
Pre-filled syringe
- SO:
-
Silicone oil
References
Singh SK, Afonina N, Awwad M, Bechtold-Peters K, Blue JT, Chou D, et al. An industry perspective on the monitoring of subvisible particles as a quality attribute for protein therapeutics. J Pharm Sci. 2010;99(8):3302–21.
Carpenter JF, Randolph TW, Jiskoot W, Crommelin DJ, Middaugh CR, Winter G, et al. Overlooking subvisible particles in therapeutic protein products: gaps that may compromise product quality. J Pharm Sci. 2009;98(4):1201–5.
Rosenberg A. Effects of protein aggregates: an immunologic perspective. AAPS J. 2006;8(3):E501–7.
Narhi LO, Jiang YJ, Cao S, Benedek K, Shnek D. A critical review of analytical methods for subvisible and visible particles. Curr Pharm Biotechnol. 2009;10(4):373–81.
Zölls S, Tantipolphan R, Wiggenhorn M, Winter G, Jiskoot W, Friess W, et al. Particles in therapeutic protein formulations, part 1: overview of analytical methods. J Pharm Sci. 2012;101(3):914–35.
Patel AR, Lau D, Liu J. Quantification and characterization of micrometer and submicrometer subvisible particles in protein therapeutics by use of a suspended microchannel resonator. Anal Chem. 2012;84(15):6833–40.
Weinbuch D, Zölls S, Wiggenhorn M, Friess W, Winter G, Jiskoot W, et al. Micro–flow imaging and resonant mass measurement (archimedes) – complementary methods to quantitatively differentiate protein particles and silicone oil droplets. J Pharm Sci. 2013;102(7):2152–65.
Sharma D, King D, Oma P, Merchant C. Micro-flow imaging: flow microscopy applied to sub-visible particulate analysis in protein formulations. AAPS J. 2010;12(3):455–64.
Demeule B, Messick S, Shire SJ, Liu J. Characterization of particles in protein solutions: reaching the limits of current technologies. AAPS J. 2010;12(4):708–15.
Zölls S, Weinbuch D, Wiggenhorn M, Winter G, Friess W, Jiskoot W, et al. Flow imaging microscopy for protein particle analysis—a comparative evaluation of four different analytical instruments. AAPS J. 2013;15(4):1200–11.
Strehl R, Rombach-Riegraf V, Diez M, Egodage K, Bluemel M, Jeschke M, et al. Discrimination between silicone oil droplets and protein aggregates in biopharmaceuticals: a novel multiparametric image filter for sub-visible particles in microflow imaging analysis. Pharm Res. 2012;29(2):594–602.
Huang CT, Sharma D, Oma P, Krishnamurthy R. Quantitation of protein particles in parenteral solutions using micro-flow imaging. J Pharm Sci. 2009;98(9):3058–71.
Kuhn M, Johnson K. Applied predictive modeling: Springer; 2013.
Kuhn M. Contributions from Jed Wing, Steve Weston, Andre Williams, Chris Keefer, Allan Engelhardt, Tony Cooper, Zachary Mayer, Brenton Kenkel, the R Core Team, Michael Benesty, Reynald Lescarbeau, Andrew Ziem and Luca Scrucca. caret: Classification and Regression Training. R package http://CRAN.R-project.org/package=caret. 2015.
Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28(5):26.
Maimon O, Rokach L. Data mining with decision trees: theory and applications. USA: World Scientific Publishing; 2012.
Breiman L, Friedman J, Stone CJ, Olshen RA. Classification and Regression Trees: Taylor & Francis; 1984.
Dago KT, Luthringer R, Lengellé R, Rinaudo G, Macher JP. Statistical decision tree: a tool for studying pharmaco-EEG effects of CNS-active drugs. Neuropsychobiology. 1994;29(2):91–6.
Bowser-Chao D, Dzialo DL. Comparison of the use of binary decision trees and neural networks in top-quark detection. Phys Rev D. 1993;47(5):1900–5.
Salzberg S. Locating protein coding regions in human DNA using a decision tree algorithm. J Comp Biol. 1995;2(3):473–85.
Kokol P, Mernik M, Završnik J, Kancler K, Malčić I. Decision trees based on automatic learning and their use in cardiology. J Med Syst. 1994;18(4):201–6.
Falconer JA, Naughton BJ, Dunlop DD, Roth EJ, Strasser DC, Sinacore JM. Predicting stroke inpatient rehabilitation outcome using a classification tree approach. Arch Phys Med Rehabil. 1994;75(6):619–25.
Breiman L. Random forests. Mach Learn. 2001;45(1):5–32.
Oshiro TM, Perez PS, Baranauskas JA. How many trees in a random forest? In: Perner P, editor. Machine learning and data mining in pattern recognition: 8th international conference, MLDM 2012, Berlin, Germany, July 13–20, 2012 proceedings. Berlin: Springer Berlin Heidelberg; 2012. p. 154–68.
Forman G. Counting positives accurately despite inaccurate classification. Machine Learning: ECML 2005: Springer; 2005. p. 564–75.
Milli L, Monreale A, Rossetti G, Giannotti F, Pedreschi D, Sebastiani F, editors. Quantification trees. Data Mining (ICDM), 2013 I.E. 13th International Conference on; 2013: IEEE.
Zölls S, Gregoritza M, Tantipolphan R, Wiggenhorn M, Winter G, Friess W, et al. How subvisible particles become invisible—relevance of the refractive index for protein particle analysis. J Pharm Sci. 2013;102(5):1434–46.
Ripple D, Hu Z. Correcting the relative bias of light obscuration and flow imaging particle counters. Pharm Res. 2015;1–20.
Joubert MK, Luo Q, Nashed-Samuel Y, Wypych J, Narhi LO. Classification and characterization of therapeutic antibody aggregates. J Biol Chem. 2011;286(28):25118–33.
ACKNOWLEDGMENTS AND DISCLOSURES
The authors would like to acknowledge Greg Downing, Mark Hu and Thomas Scherer for providing samples and valuable discussions. Daniel Coleman and Barthelemy Demeule are acknowledged for helpful discussions and reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
Details about the used training sets and size distributions for all model systems. Parameter importance and counting accuracy of the FlowCam (color) data. (DOC 2104 kb)
Rights and permissions
About this article
Cite this article
Saggu, M., Patel, A.R. & Koulis, T. A Random Forest Approach for Counting Silicone Oil Droplets and Protein Particles in Antibody Formulations Using Flow Microscopy. Pharm Res 34, 479–491 (2017). https://doi.org/10.1007/s11095-016-2079-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-016-2079-x