Abstract
COVID-19 has caused an epidemic in the entire world and it is caused by the novel virus SARS-COV-2. In severe conditions, this virus can cause a critical lung infection or viral pneumonia. To administer the correct treatment to patients, COVID-19 testing is important for diagnosing and determining patients who are infected with COVID-19, as opposed to those infected with other bacterial or viral infections. In this paper, a CResNeXt chest radiograph COVID-19 prediction model is proposed using residual network architecture. The advantage of the proposed model is that it requires lesser free hyper-parameters as compared to other residual networks. In addition, the training time per epochs of the model is very less compared to VGG19, ResNet-50, ResNeXt. The proposed CResNeXt model’s binary classification (COVID-19 versus No-Finding) accuracy is observed to be 98.63% and 99.99% and multi-class classification (COVID-19, Pneumonia, and No-Finding) accuracy is observed to be 97.42% and 99.27% on the original and augmented datasets, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Coronavirus 2019 or COVID-19 has caused a worldwide epidemic [29, 55]. This disease causes upper respiratory tract illness, such as cold, cough, fever, respiratory problems and can worsen into pneumonia or severe lung infection and inflammation. In severe cases, patients suffering from the COVID-19 infection have breathing problems and shortness of breath, requiring them to be put on ventilators in hospitals. The COVID-19 virus can cause extreme cell damage across the tissue lining of the lung air sacs. This can make breathing very difficult for the patient. In extreme cases, the body’s extreme reaction to the infection can lead to sepsis or septicemia, which may severely damage organs, cause organ failure, and cause death [30].
SARS-CoV-2 is the virus responsible for COVID-19 [47]. The coronaviruses originate in animals and are transmitted to humans. Civet cats are considered as causing the transmission of SARS-CoV, camels are considered as a possible source causing the transmission of MERS-CoV, and bats are considered as causing the transmission of SARS-CoV-2 [4, 57]. At the beginning of July 2020, there are more than 11 million cases worldwide, out of which more than 6 million have recovered, but more than 530,000 have succumbed to death. Worldwide statistics indicate a 5% death rate and 95% of patients are able to recover or fight off the disease [49, 54]. Previously published research also indicates that males are more susceptible to COVID-19 than females [25].
One of the important tasks for doctors and nurses in hospitals and testing sites is determining if a patient is infected with COVID-19 or some other viral or bacterial infection. However, countries across the globe lack the ability to adequately test and isolate the individuals infected by COVID-19 and contain the virus. Hence, there is a need for a rapid and fast automatic COVID-19 detection system. Chest radiography is one of the common modalities available in hospitals and has been shown to be useful in detecting COVID-19 infection in patient lungs [53]. Thus, there is a critical need for the development of an efficient and accurate COVID-19 prediction system from chest radiographs (X-ray images), that is both reliable and can be feasibly installed in hospital computer systems.
Nowadays machine learning (ML) and deep convolutional neural network (CNN) methods are widely used for the diagnosis of medical image. These methods are capable to provide second opinion to health care expert in daily routine. However, in ML feature extraction is difficult task and it is based on the expertise. This handcrafted features method sometimes not provide an accurate result. On the other hand, CNN based method are capable to extract features from high dimension that can be used to developed rapid and more robust system. The limitation of this CNN based method is large volume of data and architecture of model. Due to inappropriate architecture of CNN model overfitting problem may arise.
Considering above challenges we proposed residual neural network based deep CNN model CResNeXt. This model can discriminate binary (COVID-19 vs. Healthy) and Multi (COVID-19 vs. Healthy vs. Pneumonia) class classification with high accuracy. We have notice that the training and validation loss of the model is close to zero. In addition, the training time per epochs of the CResNeXt is less compare to VGG16, ResNet-50, and ResNeXt. The training time of DarkCovidNet is14 ms (milliseconds) per epochs on 625 chest X-ray images while the proposed model took 18 ms on 1756 chest X-ray images. However, proposed model has higher accuracy in binary and multi-class classification for both the original and augmented chest radiograph datasets than the aforementioned existing state-of-the-art methods.
Rest of the paper is organizes as follows, in section 2. We have discussed research papers related to COVID-19 diagnosis, Whereas, Section 3. Describe the detail architecture of proposed model. Section 4 describe the details of experimentation for binary and multi-class classification. Section 5. Gives observation of outcomes by health care expert. Finally, in section 6, performance comparison of state-of-arts deep CNN model for COVID-19 has been discussed and conclusion is presented.
2 Related works
In recent days, several deep CNN models have been developed for COVID-19 prediction from chest radiographs. In this regards, Ucar and Korkmaz [48] proposed SqueezeNet a different deep learning model for the diagnosis of COVID-19 from radiographs by classifying the radiographs into multi-class (Health, Pneumonia, and COVID-19). They tuned their SqueezeNet network using a Bayesian optimization technique. Finally, their model gained an accuracy of 76.37% on the raw dataset and 98.26% on the augmented dataset. In other research, Alom et al. [3] employed an inception residual recurrent convolutional neural network with transfer learning method named as COVID_MTNet, for the detection of the COVID-19 from X-ray and CT scan images. Their COVID-19 detection model shows an accuracy of 87% and 98.78% for X-ray and CT scan images, respectively.
Some researchers also leveraged pre-trained deep learning models for the diagnosis of COVID-19 from chest radiographs. For example, Basu et al. [8] applied a Domain Extension Transfer Learning (DETL) approach with a pre-trained deep learning network to classify the chest radiograph into a COVID-19, Normal, Pneumonia, and other diseases categories. They reported their model classification accuracy to be around 95%. Shoeibi et al. [44] summarize the ML and deep CNN model that have been used for COVID-19 diagnosis. They conluded that the these methods are capable of performing diagnosis with high efficiency. However, performance of these methods on small dataset can be further improve by designing a model based on ML and deep CNN methods. For multi-class classification high performance model still a challenge. We have summarize other research that have been conducted on COVID-19 shown in Table 1.
3 Proposed method
3.1 Dataset
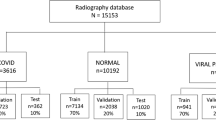
In the proposed study, we have created two different datasets (DS1 and DS2). The DS1 dataset contains COVID-19 patient images collected from the open-source repository [13]. In particular, it contains 182 radiographs of patients affected by COVID-19. Most of the chest radiographs belong to patients that fall into an age group of 12 to 83 years old, whereas some of the images were missing the age information. Moreover, the DS1 contains chest radiographs of patients who were either diagnosed as No-Finding (Health) or Pneumonia (non-COVID-19). The chest radiographs for groups No-Finding and Pneumonia were collected following the steps described in the research [26, 52]. The label for each image is defined in Data_Entry_2017.csv. The Pneumonia image of the PA view is extracted from the dataset using Python. These images are in .png format of patients 4 to 83 years and the median age is 52 years. In the proposed work, we used 1575 No-Findings and 1460 Pneumonia radiographs. Further, we constructed DS2 by augmenting images present in dataset DS1. Specifically, we utilized the Image Generator package defined in the Keras library to augment the image. In our implementation, we have applied Rotation (30ο), Horizontal-Flip, Vertical-Flip, Shear (40%), and Zoom (30%) to augment the images. After image augmentation, the dataset DS2 consisted of a total of 60,000 images (20,000 COVID-19, 20,000 Pneumonia (virus), and 20,000 No-Findings). Similar data augmentation techniques have been successfully applied in the past to generate new samples for the training of the model [33].
3.2 Proposed CResNeXt model
Medical image analysis and diagnosis is a challenging task as it requires domain knowledge and expertise in the field. However, the evolution and advancement of machine learning technologies have greatly reduced the complexity of analyzing medical images. Various machine learning techniques such as SVM (Support Vector Machine), Linear Regression, KNN (K-Nearest Neighbor), Random Forest, Naïve Bayes, etc. have helped in analyzing medical images. Nevertheless, the major concern while applying such a machine learning technique is manual feature extraction-based training and classification. On the flip side, there exist computer vision techniques, which can dramatically reduce the complexity of medical image analysis and diagnosis as they do not require manual curation of image features. Particularly, in recent years, deep learning techniques have gained widespread popularity due to their high performance and ability to extract features automatically from images without manual intervention. In the recent past, several deep learning models such as InceptionNet, GoogleNet, MobileNet, VGG16, VGG19, etc. have been used for the diagnosis of human disease [23, 35].
Highly motivated by the widespread application of deep learning techniques, in this study, we have proposed a CResNeXt network for the diagnosis of COVID-19 from a radiograph. The architecture of the proposed model is inspired by the ResNeXt model [56], which was a runner up of the ImageNet ILSVRC 2016 competition. Our motivation to extend ResNeXt for COVID-19 diagnosis is based on a fact that ResNeXt requires a relatively small number of free hyper-parameters compared to other versions of the residual network. A relatively low number of free hyper-parameters of ResNeXt model makes it a suitable choice for our COVID-19 radiograph prognosis.
The pixels’ information on radiographs is crucial for the diagnosis of diseases, because pixels loss may affect the diagnosis results. To minimize the pixel’s loss, we changed the filter size, activation function, and convolution layers’ size of the original ResNeXt model. Further, the original ResNeXt model consists of a total of 23 × 106 parameters. However, such a large number of parameters may reduce the efficiency and performance of the COVID-19 classification task. Therefore, we modified the topology of the original ResNeXt model that serves our goal of classifying COVID-19 from images.
The first convolutional layer size is changed from 64 to 32. Likewise, the kernel size is also reduced from 7 × 7 to 3 × 3. Further, the size of the other convolutional layers is also changed. However, the last convolutional layer size is retained as in the original ResNeXt model. In this way number of parameters are reduced from 23 × 106 to 18 × 106. A detailed comparison of the architecture of the ResNeXt and CResNeXt is shown in Table 2.
The major components of the proposed deep learning architecture, CResNeXt, include aggregated transformed convolutional layers, residual blocks, activation functions, and softmax optimizer at the flattened layer. The COVID-19 study model proposed in this work is shown in Fig. 1.
In ANN (Artificial Neural Network) the output of the neurons is calculated by the inner dot product of neurons weight (w) and inputs to the neurons(x) as shown in Eq. (1):
In Eq. (1), x = (x1, x2, x3……xN) and w = (w1, w2, w3. . ……wN).
Taking into account this elementary transformation we design a network of neurons. In the network of neurons, the dimension of depth is reduced by a more generic function along a new dimension. The new dimension of the proposed model is achieved by changing the elementary transformation (wkxk) of simple neurons to the aggregated transformation, as shown in Eq. (2):
where, Τk(x) represents an arbitrary function. Equivalent to a simple neuron, Τk should project N channel input vector xinto an optionally low dimensional feature vector. In a deep learning model vanishing gradient problem may occur by increasing the depth of the model. This problem is resolved by shortcut connection or residual block in our model. The shortcut connection is added after each convolution layer in the CResNeXt model see Fig. 1. The aggregated transformation in Eq. (2) serves as the residual function and output of the model is calculated as shown in Eq. (3)
The training of the data in a deep learning model is a difficult task. The weight of the layers is updated and parameters of the previous layers also change. This degrades the training performance of the model [22]. Therefore, batch normalization is used in the model. After batch normalization, Leaky Relu activation is used. This activation removes the gradient decent problem and avoids saturation of the model [21, 50]. The Leaky Relu is defined in Eq. (4):
In Eq. (4), x= input to the layers and α= gradient constant.
In a deep learning model, features are extracted by the convolutional layers. The classification of the model is predicted by the feature map extracted from the last layer by feeding to the flatten layer [17, 52, 58]. The flatten layers may get overfitted and the classification performance of the model can’t be generalized. Hinton et al. [22] suggested the concept of dropout value that can be assigned to convolution layers. This value improves the performance of the model by reducing the overfitting risk [17]. The traditional fully connected layer is replaced by the global average pooling in the ResNeXt model.
The global average pooling is used to map the features of each class with its corresponding categories. The average features map of each class is feed to the softmax layers to generate probability occurrences of class value without any optimizing parameters. In this way, the overfitting of the model is avoided. The probability of occurrences at the softmax layer is calculated by an input vector α with their feature map set β. The number of classes in the proposed work is determined by setting δ = 3 for multiclass and δ = 2 for binary class COVID-19 diagnosis. The label for binary and multiclass is assigned in k. Eq. (5) is used to define softmax optimizer for the COVID-19 diagnosis:
The cardinality C = 32 of the original model is retained in the proposed CResNeXt model. The number of the parameters in the CResNeXt model is 18 × 106 compared to 23 × 106 present in the original ResNeXt model.
4 Results
In this section, we first present the results of the proposed CResNeXt model for binary and multi-class classification of COVID-19 from radiographs on the original dataset, DS1. Then, we report the performance of CResNeXt model for binary and multi-class classification of COVID-19 on the augmented dataset, DS2.
The classification performance of the model is evaluated by the parameters precision (Pre), Recall (Re), F1-S core and Accuracy (Acc) from the confusion matrix. The indictors TP (True Positive), FP (False Positive), FN (False Negative) and TN (True Negative), calculate these measures as shown in Table 3.
4.1 Performance of CResNeXt on the original dataset, DS1
In this section, we have presented the results of the experiments we performed to identify COVID-19 from the original radiograph dataset, DS1.
4.1.1 Binary class classification
First, the proposed CResNeXt deep learning model is trained to classify radiographs into two categories: COVID-19 and No-Findings. Then, the performance of the CResNeXt binary class classifier is estimated using 5-fold cross-validation (CV), an evaluation method that is simple to use and helpful to construct a bias-free model [9, 14, 20, 38]. In the 5-fold cross-validation approach, cross-validation is performed in 5 folds, where the data is segmented into 5 parts, which are each of about the same size. During each fold, an individual part is set aside for validation and the rest of the parts are used to train the classifier. This process is repeated 5 times so that each part is set aside once for validation. Consequently, the 5 estimates of accuracies obtained from 5 folds of validation are combined to find the average [20].
To perform 5-fold cross-validation of binary classification model, we have used a dataset consisting of 182 COVID-19 and 1575 No-Finding radiographs from dataset DS1, where 80% of the radiographs are used for training and 20% for validation. The images are resized to 256 × 256 pixels before feeding to the deep learning model. We trained CResNeXt for 100 epochs with a batch size of 15 and an initial learning rate of 3e−3. The confusion matrix (CM) obtained for each fold of the 5-fold cross-validation, performed on the CResNeXt binary class classifier, is shown in Fig. 2. It can be noted from the confusion matrix that the CResNeXt is able to classify COVID-19 vs. No-Finding with an accuracy of more than 98% for 4 out of 5 folds (see Fig. 2 (a), (c), (d), and (e)) and more than 95% for the remaining fold (see Fig. 2 (b)). Further, Table 4 presents individual folds and average sensitivity, specificity, precision, recall, F1-score, and accuracy of the binary classification model. From Table 2, we observe that the CResNeXt model achieves an average binary classification sensitivity, specificity, precision, recall, F1-score, and accuracy of 99.24%, 99.73%, 99.24%, 99.23%, 99.23%, and 98.63%, respectively. These results show that the proposed CResNeXt binary classifier can classify COVID-19 and No-Finding with a significantly high accuracy of 98.63%.
Moreover, Fig. 3 illustrates the training and validation loss as well as accuracy graphs of the binary class classification model. It is evident from Fig. 3 (a) that the training and validation loss of the model gets close to zero and saturates after 70 epochs. Likewise, from Fig. 3 (b), we can note that the training and validation accuracy of the model gets close to 1 and saturates after 70 epochs.
4.1.2 Multi-class classification
In addition to binary classification, the CResNeXt deep learning model is trained to classify radiographs into three categories: COVID-19, No-Findings, and Pneumonia. We used 5-fold cross-validation (CV) approach to evaluate the performance of the CResNeXt multi-class classifier. To perform 5-fold CV, we used a dataset consisting of 182 COVID-19, 1575 No-Finding, and 1460 Pneumonia radiographs in dataset DS1, where 80% of the radiographs are used for training and 20% for validation. The images in all three classes are resized to 256 × 256 pixels before feeding to the deep learning model. The proposed CResNeXt model is trained for 100 epochs with a batch size of 15 and an initial learning rate of 3e−3.
Figure 4 shows the CM obtained for each fold of the 5-fold CV, performed on the CResNeXt multi-class classifier. It is evident from the CM that the CResNeXt is able to classify COVID-19, No-Finding, and Pneumonia with an accuracy of more than 97% for 4 out of 5 folds (see Fig. 4 (b), (c), (d), and (e)) and more than 89% for the remaining fold (see Fig. 4 (a)). Additionally, Table 5 highlights that the CResNeXt model achieves an average multi-class classification sensitivity, specificity, precision, recall, F1-score, and accuracy of 98.45%, 96.50%, 97.63%, 97.57%, 97.6%, and 97.42%, respectively. These results highlight that the proposed CResNeXt multi-class classifier can classify COVID-19, No-Finding, and Pneumonia with significantly high accuracy of 97.42%.
Furthermore, Fig. 5 presents the training and validation loss as well as accuracy graphs of the multi-class classification model. It can be noted from Fig. 5 (a) that the training loss of the model is significantly low since the beginning of the training process and the validation loss remains less than 0.0002 throughout the validation process. Likewise, it is evident from Fig. 5 (b) that the training and validation accuracy of the model gets close to 100% and saturates after 35 epochs.
4.2 Performance of CResNeXt on augmented dataset, DS2
Deep learning algorithms often leverage the image augmentation technique for improved performance. Motivated by the success of image augmentation techniques in deep learning applications, we adopted different image augmentation techniques to increase the size of our dataset and consequently, trained and validated the proposed approach. Here, we present the results of the experiments we performed to identify COVID-19 from the augmented radiograph dataset, DS2.
4.2.1 Binary class classification
To assess the binary class classification performance of CResNeXt model on augmented DS2, we adopted a simple model training and independent testing approach. The dataset consisting of 20,000 COVID-19 and an equal amount of No-Finding images on dataset DS2 is randomly divided into 80% training and 20% testing. After training the model with 80% of images from both the classes (COVID-19 and No-Finding) for 100 epochs with a batch size of 15, the model is tested with the remaining 20% of the independent test dataset. None of the images in the independent test set was used during the training of the model. The CM of the binary class classification model on the augmented independent test dataset is shown in Fig. 6. The model is tested on the independent testing image of 4000 for each class. The CM shows that the proposed model is highly sensitive towards accurately detecting COVID-19. Specifically, the model is able to accurately predict 3999 out of 4000 total COVID-19 radiographs present in the augmented independent test dataset.
Additionally, Table 6 illustrates the details of different performance metrics obtained while independently testing the model trained on the augmented dataset. In Table 6, we observe that the CResNeXt model achieves a binary classification sensitivity, specificity, precision, recall, F1-score, and accuracy of 100%, 99.98%, 100%, 99.98%, 99.99%, and 99.98%, respectively for COVID-19. These results show that the proposed CResNeXt binary classifier can classify COVID-19 and No-Finding with significantly high accuracy of 99.99%. These results indicate that the proposed CResNeXt binary classifier is highly accurate and can be utilized by health experts to assist them in their day to day work.
Furthermore, we present the training and validation loss, as well as accuracy graphs of the binary class classification model on dataset DS2 in Fig. 7. From Fig. 7 (a), we see that the training loss starts with a value of 0.04 and validation loss starts with a value of 0.09. Initially, validation loss fluctuates for some epochs and starts showing no major change in the loss value after 60 epochs as well as the loss reduced to a value close to 0. A similar trend can be seen in the training and validation accuracy shown in Fig. 7 (b). The training accuracy starts with a value of 98% and a validation accuracy state with a value of 97%. The classification accuracy fluctuates for some epochs. After 60 epochs, model classification accuracy reaches close to 100 for both training and validation.
4.2.2 Multi-class classification
Here, we assess the multi-class classification performance of the CResNeXt model on augmented dataset DS2 through model training followed by independent testing. Our augmented dataset consists of 20,000 COVID-19 and an equal number of No-Finding and Pneumonia images. In our implementation, we used 80% of the images for training and the remaining 20% images for testing our multi-class classification model. The training of the model is performed for 100 epochs with a batch size of 15. None of the images used for the independent testing was utilized for the training of the model. The CM of our multi-class classification model on the augmented independent test dataset is shown in Fig. 8. The CM shows that the proposed multi-class model is highly sensitive towards accurately detecting COVID-19. Specifically, the model is able to accurately predict 3999 out of 4000 total COVID-19 radiographs present in the augmented independent test dataset.
Furthermore, in Table 7 we present the details of different performance metrics obtained while independently testing the model trained on the dataset DS2. From Table 5, it is evident that the proposed model attains an average sensitivity, specificity, precision, recall, F1-score, and accuracy of 98.33%, 98%, 99.33%, 99.33%, 99.33%, and 99.27%, respectively for classifying COVID-19 from No-Finding, and Pneumonia. Additionally, the individual class accuracy of the model for detecting COVID-19, No-Finding, and Pneumonia are 99.98%, 99.78%, and 98.07%, respectively. These results indicate that the proposed model is able to classify COVID-19, No-Finding, and Pneumonia with higher accuracy and therefore, can assist health care experts in detecting COVID-19 from chest radiographs.
Moreover, in Fig. 9, we present the training and validation loss as well as accuracy graphs of the multi-class classification model on the augmented dataset DS2. The loss graphs in Fig. 9 (a) shows that the training loss remains lower than 0.5 throughout the training process and settles at close to zero towards the end of the training of the model whereas, the validation loss fluctuates heavily at the beginning and settles at close to zero towards the end of the training of the model. A similar trend is seen in Fig. 9 (b), which shows that the training accuracy remains above 0.9 and gets closer to 1 towards the end of the training of the model whereas, the validation accuracy fluctuates significantly initially and gets closer to 1 towards the end of the training and validation of the multi-class model on DS2. These results indicate that the proposed model yields consistently better performance for both binary and multi-class classification of chest radiographs for both original and augmented datasets. Thus, the proposed model can be trusted by the health experts and can be utilized to assist them in diagnosing COVID-19 from chest radiographs. The summary of performance measures for DS1 and DS2 is shown in Table 8.
4.3 Proposed 2Scale deep CNN model
In the present study, we have applied texture feature and deep feature fusion for the diagnosis of COVID-19. In this approach, two-scale deep CNN architecture has been developed, for both the scale, AlexNet has been used. The detail architecture of the method is shown in Fig. 10.
AlexNet is a sequential model having 8 layers. In which 5 are convolution layer and with a combination of max-pooling layers. After that, it has 3 fully connected layers. In all the layers Relu activation function has been used. The input to the model is original image and their corresponding LBP (Local Binary pattern) images. The LBP feature find texture pattern in an image that can be used for the classification of an image.
The input to the model is original image and their corresponding LBP image of size 224x224x3 pixel and training and validation is performed on the DS1 and DS2 Under same condition as discussed in section 3. The training and validation loss and accuracy is shown in Fig. 11(a) and (b) respective. We can observe from 11(a) training and validation loss reaches close to zero after 80 epochs. Similarly, in Fig. 11(b) training accuracy reaches 100% after 80 epochs.
4.4 Evaluation of model outcomes by the healthcare experts
Here, we present the interpretation of the results of the CResNeXt model by an expert radiologist and a physician. The proposed CResNeXt model automatically detects COVID-19 from chest radiographs, without the need for manual curation of the features. This model can be extremely useful as it provides a second opinion to healthcare experts and supports them in their day-to-day clinical decisions related to COVID-19. This model can support healthcare experts make a quick (the diagnosis process based on the proposed model is fast) and accurate diagnosis in their routine work. To assess the robustness of the proposed model, we have shared the outcomes of the model with expert radiologists and physicians. Specifically, we provided them with the images that were misclassified by the model and the actual labels. The radiologist and physician perspectives on the output of CResNeXt model are as follows:
-
The CResNeXt model performed significantly well in detecting COVID-19 cases for the binary classification task on both original and augmented datasets.
-
For the multi-class classification task, where Pneumonia images are also included in the training and testing, the model diagnosed some of the patients with Pneumonia as COVID-19 (see Fig. 12 (a)). The hazy opacities in the chest radiograph can be seen both in pneumonia (bacterial) and in the case of lung involvement from COVID-19, that is why the patient identified with Pneumonia is diagnosed as COVID-19 by the model. Particularly, in Fig. 12 (a), patchy opacities seen in medial right upper and lower lobe and right perihilar region and left upper lobe with air bronchogram at places favor pneumonia.
-
As the model is sensitive towards detecting Pneumonia and COVID-19, it could diagnose some of the images as Pneumonia (see Fig. 12 (b)) or COVID-19 (see Fig. 12 (c)) even though they are marked as no-finding in the dataset. The reason model could do so is that in certain radiographs, the image has poor resolution, has image artifacts, or has some mass or growth in the lung tissue.
-
The model sometimes incorrectly diagnoses the patient as No-Finding, when in fact the patient is marked as COVID-19 or Pneumonia in the dataset. Figure 12 (d) and (e) shows an example of the patient diagnosed as No-Finding whereas, they were marked as COVID-19 and pneumonia in the database, respectively. This is due to the fact that some pixels in the lung regions in the image have intensities in a similar range as regions outside of the lung. Particularly, in Fig. 12 (d) multifocal atelectasis is seen in the lower aspect of bilateral upper lobes, middle lobe and bilateral lower lobes predominantly in the peripheral region, however, no signs of pleural effusion are evident. Likewise, Fig. 12 (e) has hazy opacities in the medial aspect of the right middle lobe with pleural effusion favoring pneumonia.
-
None of the images marked as COVID-19 were predicted as Pneumonia by the model
Images evaluated by the radiologist, physician and CResNeXt model. (a) predicted as COVID-19 by model but actual class is Pneumonia, (b) and (c) predicted as Pneumonia and COVID-19, respectively by model but actual class is No-Finding, and (d) and (e) predicted as No-Finding by the model but actual class is COVID-19 and Pneumonia, respectively
Further, Fig. 13 shows the heatmap of the chest radiographs of COVID-19 patient that are correctly and misclassified by the CResNeXt model. Moreover, Fig. 14 shows the disparities seen between a few COVID-19 and Pneumonia case images and the radiologist and physicians’ findings and impression.
5 Discussion
Most of the studies discussed in this section utilize a computational/automated approach for the diagnosis of COVID-19 from chest radiographs. These approaches either utilize the COVID-19 image dataset available from the GitHub repository [13] or [12]. In the recent past, Wang et al. [50] developed a COVID-NET model to classify chest radiographs into three classes: Normal, Pneumonia, and COVID-19. To train their model they used a dataset, namely COVIDx, which is created leveraging data augmentation techniques. In addition to COVID-NET, they explored some other deep learning techniques such as VGG19 and Resnet-50 for multi-class classification of radiographs. Through a comprehensive analysis, they concluded that their COVID-NET model performs better compared to VGG19 and Resnet-50 models. As reported, their model results in an accuracy of 95% for Normal, 94% for Pneumonia, and 91% for COVID-19 classes on the augmented dataset.
Alternatively, Apostolopoulos and Mpesiana [5] adopted a transfer learning approach for the classification of radiographs into two classes: COVID-19 and Normal as well as three classes: COVID-19, Normal, and Pneumonia. Their method explores deep learning models including VGG19, Inception, MobileNet v2, Xception, and Inception ResNet v2 along with transfer learning techniques for automatic detection of COVID-19 from a radiograph. As reported, among all the models explored in their study, MobileNet v2 attains the highest binary and multi-class classification accuracy of 96.78% and 94.72%, respectively.
Another approach, proposed by Hemdan et al. [21] utilize a deep learning technique, namely COVIDX-Net to diagnose COVID-19 in radiographs. They used 50 chest radiographs with 25 confirmed positive COVID-19 cases and 25 normal images. Their model achieved an accuracy of 90% for binary class classification with 25 images in each class. Luz et al. [31] designed EfficientNet B3, a deep learning-based model, for the classification of radiographs into three categories: COVID-19, Pneumonia, and Healthy. EfficientNet B3 achieved an overall accuracy of 93.9%.
Ying et al. [46] applied DRE-Net for the diagnosis of COVID-19 and Pneumonia CT scan images. The DRE-NET model is built on the pre-trained ResNet50 model. Their model achieved an 86% success rate in discriminating COVID-19 and bacterial Pneumonia from CT images. Sethy and Behera [42] utilized Support Vector Machine (SVM) and ResNet50 to detect COVID-19 infected patients using chest radiographs. Particularly, SVM is used to classify the deep features obtained from images by applying the ResNet50 deep learning model. Their combined Resnet-50 and SVM model achieved an accuracy of 95.38%. Wang et al. [51] designed an inception migration-learning algorithm to diagnose COVID-19 using CT scan images. As stated, their model achieved an accuracy of 82.9%. Khan et al. [27] developed CoroNet, a deep learning model to automatically detect COVID-19 infection from chest radiographs. However, CoroNet was trained and tested on a small dataset to classify radiographs into binary, 3-class and 4-class individually. Their model attains a binary, 3-class, and 4-class classification accuracy of 99%, 95%, and 89.6%, respectively.
Pereira et al. [40] perform a diagnosis of Pneumonia, Healthy, and COVID-19 using radiographs. Their method uses multi-class classification and a hierarchical classification approach. For multi-class classification, they explore individual machine learning methods such as k-Nearest Neighbor (kNN), SVM, Multilayer Perceptrons (MLP), Decision Trees (DT), and Random Forests (RF). As well, for hierarchical classification, they utilized the Clus-HMC framework. In addition, the authors balanced the dataset using resembling techniques. Finally, their method achieved F1-score of 0.65 for the multi-class approach and a F1-score of 0.89 for COVID-19 identification in a hierarchical scenario.
Oh et al. [37] proposed a patch-based deep learning technique for detecting COVID-19 from radiographs, which involved applying a segmentation network and classification network in consecutive stages. The segmentation network is used to extract lung areas. These lung areas are then used for training the individual deep learning models (ResNet-18), after which the final decision is made based on majority voting. Their patch-based deep learning technique resulted in COVID-19 detection accuracy of 88.9%. Rahimzadeh and Attar [41] concatenate two models Xception and ResNet50V2 for the diagnosis of the COVID-19. The authors apply data augmentation techniques to increase the size as well as construct a balanced dataset. Their model achieved a classification accuracy of 91.4% for multi-class (Normal, Pneumonia, and COVID-19) classification. Pathak et al. [39] adopted ResNet-50 along with deep transfer learning to classify COVID-19 patients using CT scan images. Their model achieved an accuracy of 93.02% in detecting COVID-19 from CT scan images.
In summary, the methods discussed above applied various techniques for automated detection of COVID-19 including, individual machine learning methods, deep learning-based approaches, transfer learning techniques and a combination of these. Moreover, some of these methods diagnosed COVID-19 from chest radiographs whereas, others utilized CT scan images. Overall, these methods have notably advanced the study of COVID-19. However, the results indicate that the performance of the above-listed methods is limited and possible avenues to improve the performance of the automated diagnosis of COVID-19 should be explored. The COVID-19 disease is evolving day by day and as a result, the COVID-19 pandemic is still around the corner. Scientists and health experts deal with new challenges every day. Therefore, it is now, more than ever, important to advance the study of COVID-19 disease.
In this work, we propose a deep learning-based method, called CResNeXt for automatic detection of COVID-19 infection from chest radiographs. The proposed model is an extension of ResNeXt architecture. It is trained with a batch size of 15 for 100 epochs and an initial learning rate of 3e−3. The model is feed with an input image of size 256x256x3. Further, it is validated using 5-fold cross-validation on the original dataset, DS1 as well as trained and independently testing on the augmented dataset, DS2. The DS1 dataset consist 3217 radiographs, where 182 images belong to COVID-19, 1460 images belong to Pneumonia (virus), and 1575 images belong to the No-Finding categories. On the other hand, the DS2 dataset consists of a total of 60,000 images, where each category COVID-19, Pneumonia, and No-Finding contains 20,000 images each. The classification accuracy of the proposed CResNeXt model on dataset DS1 for binary class (COVID-19 and No-Finding) and multi-class (COVID-19, Pneumonia, and No-Finding) is 98.63% and 97.42%, respectively. Moreover, the classification accuracy of CResNeXt on dataset DS2 for the binary class and multi-class is 100% and 99.27%, respectively.
Tables 9 and 10 illustrates the performance comparison of the proposed model with the existing state-of-the-art methods on datasets DS1 and DS2, respectively. From Table 9, we can see that the CoroNet model proposed by Khan et al. results in the highest binary class classification accuracy of 99% on the DS1 dataset. Whereas, the proposed CResNeXt model attains binary class accuracy of 98.63% on the DS1 dataset, which is very close to the accuracy of the CoroNet model. On the flip side, the proposed CResNeXt model achieves the highest multi-class classification accuracy of 97.42% on the original dataset compared to other existing methods shown in Table 6. The second best method that achieves an accuracy of 93.48% for multi-class classification on the original dataset is VGG19, which was proposed by Apostolopoulos et al. [5]. Moreover, from Table 7, we can see that the proposed CResNeXt model results in the highest binary and multi-class classification accuracy of 99.99% and 99.27% on the augmented dataset, respectively compared to the existing methods trained and tested on the augmented dataset.
Moreover, we have used the area under the curve (AUC) and receiver operating characteristic (ROC) curve measures to further evaluate the performance of the proposed model. The ROC curve gives a trade-off between TPR (True Positive Rate) and FPR (False Positive Rate) at different classification thresholds [39]. Figure 15 presents the ROC curve generated by CResNeXt for multi-class and binary class classification while the predictions are evaluated on DS2. The curve in Fig. 15 highlights the strength of CResNeXt in achieving a high TPR of close to 100% at a very low FPR of close to 0% for both multi-class and binary class classification. Moreover, the micro and macro-average AUC score given by CResNeXt for multi-class classification is 0.99. Likewise, the AUC for binary class classification is 100%. The value of AUC close to 1 for both multi and binary class models indicates that the model is highly accurate in detecting COVID-19 from chest radiograph.
These results indicate that the proposed method outperforms the state-of-the-art methods and can be utilized by health experts to effectively diagnose COVID-19 infection from chest radiographs. The radiograph diagnosis is fast, reliable, and widely used for the diagnosis of COVID-19 patients. Specifically, the proposed model can be used for the initial screening of COVID-19 patients from a radiograph. After the initial screening by the model, if a patient is diagnosed with COVID-19 then the patient can be sent for the PCR test. Meanwhile, a patient can be given initial treatment without delay. However, if the patient is tested negative during initial screening then the patient may not need to go for the PCR test.
6 Conclusion
In this work, we have designed and developed two deep learning-based methods, namely CResNeX and 2Scale feature fusion model for automatic diagnosis of COVID-19 infection from chest radiographs. To improve the prediction accuracy of COVID-19 diagnosis in less training time. The proposed CResNeXt model achieves binary (COVID-19 and No-Finding) and multi-class (COVID-19, Pneumonia, and No-Finding) classification accuracy of 98.63% and 97.42% on dataset DS1, respectively. Further, the CResNeXt results in binary and multi-class classification accuracy of 99.99% and 99.27% on dataset DS2, respectively. The 2Scale feature fusion model achieved an accuracy of 98.75% and 98.63% for binary and multi-class classification respectively. We have found that these models, outperforms existing methods based on both original and augmented datasets. The comparison of proposed method is shown in Table 11. These promising results indicate that CResNeXt and 2Scale feature fusion models are very robust and can be effectively used by health experts to diagnose COVID-19 from chest radiographs.
Limitation and future study
In the proposed study, we have kept external validation as our limitation. In addition, limitation of input data is another weakness. In the future work, probability value obtained from Softmax optimizer for a class will be map using temperature-scaling method. So that external validation can be, perform for highly sensitive model. Texture features like LBP, HOG, GLCM and deep features together can be a new step towards designing a robust system.
References
Abbas A, Abdelsamea MM, Gaber MM (2020) Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network, arXiv preprint arXiv:2003.13815
Alizadehsani R, Sharifrazi D, Izadi NH, Joloudari JH, Shoeibi A, Gorriz JM, Hussain S, Arco JE, Sani ZA, Khozeimeh F, Khosravi A, Nahavandi S, Islam SMS, Acharya UR (2021) Uncertainty-aware semi-supervised method using large unlabeled and limited labeled COVID-19 data. ACM Trans Multimed Comput Commun Appl (TOMM) 17(3s):1–24
Alom MZ, Rahman M, Nasrin MS,Taha TM, Asari VK (2020) COVID_MTNet: COVID-19 Detection with Multi-Task Deep Learning Approaches, arXiv preprint arXiv:2004.03747
Alshukairi AN, Zheng J, Zhao J, Nehdi A, Baharoon SA, Layqah L, Bokhari A, Johani SMA, Samman N, Boudjelal M (2018) High prevalence of MERS-CoV infection in camel workers in Saudi Arabia. MBio 9(5):e01985-18
Apostolopoulos ID, Mpesiana TA (2020) Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med 43:1–640
Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A (2020) Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med 121:103795
Ayyar MP, Benois-Pineau J, Zemmari A (2021) A hierarchical classification system for the detection of Covid-19 from chest X-ray images. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 519–528. https://doi.org/10.1109/ICCVW54120.2021.00064
Basu S, Mitra S, Saha N (2020) Deep learning for screening covid-19 using chest x-ray images. In: 2020 IEEE Symposium Series on Computational Intelligence (SSCI). IEEE, pp 2521–2527. https://doi.org/10.1109/SSCI47803.2020.9308571
Bishop C (2009) Pattern recognition and machine learning. Springer, New York
Brunese L, Mercaldo F, Reginelli A, Santone A (2020) Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput Methods Prog Biomed 196:105608. https://doi.org/10.1016/j.cmpb.2020.105608
Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al-Emadi N ( 2020) Can AI help in screening viral and COVID-19 pneumonia?, arXiv preprint arXiv:2003.13145
Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M (2020) Covid-19 image data collection: Prospective predictions are the future, arXiv preprint arXiv:2006.11988
"COVID-19, (n.d.) " https://github.com/ieee8023/covid-chestxray-dataset?
Duda RO, Hart PE, Stork DG (2000) Pattern classification. Wiley, Hoboken
Farooq, M., & Hafeez, A. (2020). Covid-resnet: a deep learning framework for screening of covid19 from radiographs. arXiv preprint arXiv:2003.14395
Ghassemi N, Shoeibi A, Khodatars M, Heras J, Rahimi A, Zare A, ... Gorriz JM (2021) Automatic diagnosis of covid-19 from ct images using cyclegan and transfer learning. arXiv preprint arXiv:2104.11949
Goodfellow I, Warde-Farley D, Mirza M, Courville A, Bengio Y (n.d.) Maxout networks. pp. 1319–1327
Haghanifar A, Majdabadi MM, Choi Y, Deivalakshmi S, Ko S (2020) Covid-cxnet: detecting covid-19 in frontal chest x-ray images using deep learning. arXiv preprint arXiv:2006.13807
Hall LO, Paul R, Goldgof DB, Goldgof GM (2020) Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv:2004.02060. https://doi.org/10.48550/arXiv.2004.02060
Hastie T, Tibshirani R, Friedman JH, Friedman JH (2009) The elements of statistical learning: data mining, inference, and prediction. Springer, New York, vol 2, pp 1–758
Hemdan EE-D, Shouman MA, Karar ME (2020) Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images, arXiv preprint arXiv:2003.11055
Hinton GE, Srivastava N, Krizhevsky A, Sutskever I, Salakhutdinov RR (2012) Improving neural networks by preventing co-adaptation of feature detectors. arXiv preprint arXiv:1207.0580. https://doi.org/10.48550/arXiv.1207.0580
Hirano G, Nemoto M, Kimura Y, Kiyohara Y, Koga H, Yamazaki N, Christensen G, Ingvar C, Nielsen K, Nakamura A, Sota T, Nagaoka T (2020) Automatic diagnosis of melanoma using hyperspectral data and GoogLeNet. Skin Res Technol 26(6):891–897
Horry MJ, Chakraborty S, Paul M, Ulhaq A, Pradhan B, Saha M, Shukla N (2020) X-ray image based COVID-19 detection using pre-trained deep learning models
Jin J-M, Bai P, He W, Wu F, Liu X-F, Han D-M, Liu S, Yang J-K (2020) Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health 8:152
Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F (2018) Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172(5):1122–1131.e9
Khan AI, Shah JL, Bhat MM (2020) Coronet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Prog Biomed 196:105581
Khozeimeh F, Sharifrazi D, Izadi NH, Joloudari JH, Shoeibi A, Alizadehsani R, Gorriz JM, Hussain S, Sani ZA, Moosaei H, Khosravi A, Nahavandi S, Islam SMS (2021) Combining a convolutional neural network with autoencoders to predict the survival chance of COVID-19 patients. Sci Rep 11(1):1–18
Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR (2020) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents 55(3):105924. https://doi.org/10.1016/j.ijantimicag.2020.105924
Liu Y, Yan L-M, Wan L, Xiang T-X, Le A, Liu J-M, Peiris M, Poon LL, Zhang W (2020) Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 20:656–657
Luz E, Silva PL, Silva R, Moreira G (2020) Towards an efficient deep learning model for covid-19 patterns detection in x-ray images, arXiv preprint arXiv:2004.05717
Medhi K, Jamil M, Hussain MI (2020) Automatic detection of COVID-19 infection from chest x-ray using deep learning. medrxiv
Mikołajczyk A, Grochowski M (n.d.) Data augmentation for improving deep learning in image classification problem. pp. 117–122
Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ (2020) Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal 65:101794
Nahata H, Singh SP (2020) Deep learning solutions for skin cancer detection and diagnosis. Machine Learning with Health Care Perspective: Machine Learning and Healthcare 13:159–182
Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal Appl 24:1207–1220
Oh Y, Park S, Ye JC (2020) Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans Med Imaging 39:2688–2700
Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR (2020) Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med 121:103792
Pathak Y, Shukla PK, Tiwari A, Stalin S, Singh S, Shukla PK (2020) Deep transfer learning based classification model for COVID-19 disease. IRBM
Pereira RM, Bertolini D, Teixeira LO, Silla CN Jr, Costa YM (2020) COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Methods Prog Biomed 194:105532
Rahimzadeh M, Attar A (2020) A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Informatics in Medicine Unlocked 19:100360
Sethy PK, Behera SK (2020) Detection of coronavirus disease (COVID-19) based on deep features. Preprints.org. https://doi.org/10.20944/preprints202003.0300.v1
Sharifrazi D, Alizadehsani R, Roshanzamir M, Joloudari JH, Shoeibi A, Jafari M, Hussain S, Sani ZA, Hasanzadeh F, Khozeimeh F, Khosravi A, Nahavandi S, Panahiazar M, Zare A, Islam SMS, Acharya UR (2021) Fusion of convolution neural network, support vector machine and Sobel filter for accurate detection of COVID-19 patients using X-ray images. Biomed Signal Process Control 68:102622
Shoeibi A, Khodatars M, Alizadehsani R, Ghassemi N, Jafari M, Moridian P, ..., Shi P (2020) Automated detection and forecasting of covid-19 using deep learning techniques: A review. arXiv preprint arXiv:2007.10785
Silva P, Luz E, Silva G, Moreira G, Silva R, Lucio D, Menottu D (2020) Efficient deep learning model for COVID-19 detection in large CT images datasets: a cross-dataset analysis
Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R, (2020) Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images, medRxiv
Tetro JA (2020) Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect 22(2):72–73
Ucar F, Korkmaz D (2020) COVIDiagnosis-Net: Deep Bayes-SqueezeNet based Diagnostic of the Coronavirus Disease 2019 (COVID-19) from X-Ray Images. Med Hypotheses 140:109761
W. H. Organization, (2020) Coronavirus disease (COVID-19): situation report, 165
Wang L, Wong A (2020) COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images, arXiv preprint arXiv:2003.09871
Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X (2020) A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19), MedRxiv
Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2097–2106. https://doi.org/10.1109/CVPR.2017.369
Wong HYF, Lam HYS, Fong AH-T, Leung ST, Chin TW-Y, Lo CSY, Lui MM-S, Lee JCY, Chiu KW-H, Chung T (2020) Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology:201160
Worldometer. (n.d.) "COVID-19 Coronavirus Pandemic," https://www.worldometers.info/coronavirus/
Wu Z, McGoogan JM (2020) Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. Jama 323(13):1239–1242
Xie S, Girshick R, Dollár P, Tu Z, He K (n.d.) Aggregated residual transformations for deep neural networks. pp. 1492–1500
Xu J, Zhao S, Teng T, Abdalla AE, Zhu W, Xie L, Wang Y, Guo X (2020) Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 12(2):244
Zeiler MD, Fergus R (2013) Stochastic pooling for regularization of deep convolutional neural networks, arXiv preprint arXiv:1301.3557
Zhang J, Xie Y, Li Y, Shen C, Xia Y (2020) Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338, 27
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yadav, D.P., Jalal, A.S., Goyal, A. et al. COVID-19 radiograph prognosis using a deep CResNeXt network. Multimed Tools Appl 82, 36479–36505 (2023). https://doi.org/10.1007/s11042-023-14960-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14960-7